Descriptive Analysis
First created on Nov 7, 2019. Updated on Nov 09, 2019
- Step 1: Read in the BRFSS 2018 data set and check main variables
- Step 2: Variable Histograms
- Step 3: Frequency Calculations for Table 1
Step 1: Read in the BRFSS 2018 data set and check main variables
source("../code/100_Dependencies.R", echo = TRUE)
##
## > library(foreign)
##
## > library(Hmisc)
## Loading required package: lattice
## Loading required package: survival
## Loading required package: Formula
## Loading required package: ggplot2
##
## Attaching package: 'Hmisc'
## The following objects are masked from 'package:base':
##
## format.pval, units
##
## > library(e1071)
##
## Attaching package: 'e1071'
## The following object is masked from 'package:Hmisc':
##
## impute
##
## > library(gtools)
##
## Attaching package: 'gtools'
## The following object is masked from 'package:e1071':
##
## permutations
##
## > library(MASS)
##
## > library(ggplot2)
##
## > library(gridExtra)
##
## > library(broom)
##
## > library(scales)
##
## > library(ggfortify)
##
## > library(gvlma)
source("../code/200_Calculate totals.R", echo = TRUE)
##
## > BRFSS <- read.csv(file = "../output/BRFSS.csv", header = TRUE,
## + sep = ",")
##
## > nrow(BRFSS)
## [1] 266109
##
## > TeethFreq <- table(BRFSS$RMVTETH5)
##
## > TeethFreq
##
## 0 1 2 3
## 104802 90192 44317 26798
##
## > write.csv(TeethFreq, file = "../output/frequencies/teethFreq.csv")
##
## > TeethFreq/nrow(BRFSS) * 100
##
## 0 1 2 3
## 39.38311 33.89288 16.65370 10.07031
##
## > cvdFreq <- table(BRFSS$CVDINFR5)
##
## > cvdFreq
##
## 0 1
## 243070 23039
##
## > write.csv(cvdFreq, file = "../output/frequencies/cvdFreq.csv")
##
## > cvd_teethFreq <- table(BRFSS$CVDINFR5, BRFSS$RMVTETH5)
##
## > write.csv(cvd_teethFreq, file = "../output/frequencies/cvd_teethFreq.csv")
##
## > table(BRFSS$CVDINFR5, BRFSS$RMVTETH5)
##
## 0 1 2 3
## 0 99474 83224 38416 21956
## 1 5328 6968 5901 4842
##
## > sexFreq <- table(BRFSS$SEX)
##
## > write.csv(sexFreq, file = "../output/frequencies/sexFreq.csv")
##
## > table(BRFSS$RACEGRP)
##
## 0 1 2 3 4 9
## 214832 20092 13614 3541 11005 3025
Step 2: Variable Histograms
A histogram was generated for each variable in the study including:
- Reported Cardiovascular Disease - independent (outcome) variable
- Reported Teeth Removed - dependent (exposure) variable
- All studied potential confounders (Sex, Race, Age, Education, Income, Dental Visit, Diabetes, Smoker, Exercise, and BMI Category)
source("../code/205_key variable plots.R", echo = TRUE)
##
## > BRFSS <- read.csv(file = "../output/BRFSS.csv", header = TRUE,
## + sep = ",")
##
## > BRFSS$CVDINFR5 <- as.factor(BRFSS$CVDINFR5)
##
## > levels(BRFSS$CVDINFR5) <- c("Yes", "No")
##
## > BRFSS$RMVTETH5 <- as.factor(BRFSS$RMVTETH5)
##
## > levels(BRFSS$RMVTETH5) <- c("None", "1-5", "6+", "All")
##
## > BRFSS$SEX2 <- as.factor(BRFSS$SEX2)
##
## > levels(BRFSS$SEX2) <- c("Female", "Male", "NR")
##
## > BRFSS$RACEGRP <- as.factor(BRFSS$RACEGRP)
##
## > levels(BRFSS$RACEGRP) <- c("White", "Black", "Hisp",
## + "Asian", "Other", "NR")
##
## > p1 <- ggplot(BRFSS, aes(CVDINFR5, fill = CVDINFR5))
##
## > p1 <- p1 + geom_bar(color = "Black") + scale_fill_brewer(palette = "Blues") +
## + theme_minimal() + theme(axis.text.x = element_text(angle = 45)) .... [TRUNCATED]
##
## > p2 <- ggplot(BRFSS, aes(RMVTETH5, fill = RMVTETH5))
##
## > p2 <- p2 + geom_bar(color = "Black") + scale_fill_brewer(palette = "Blues") +
## + theme_minimal() + theme(axis.text.x = element_text(angle = 45)) .... [TRUNCATED]
##
## > p3 <- ggplot(BRFSS, aes(SEX2, fill = SEX2))
##
## > p3 <- p3 + geom_bar(color = "Black") + scale_fill_brewer(palette = "Blues") +
## + theme_minimal() + theme(axis.text.x = element_text(angle = 45)) .... [TRUNCATED]
##
## > p4 <- ggplot(BRFSS, aes(RACEGRP, fill = RACEGRP))
##
## > p4 <- p4 + geom_bar(color = "Black") + scale_fill_brewer(palette = "Blues") +
## + theme_minimal() + theme(axis.text.x = element_text(angle = 45)) .... [TRUNCATED]
##
## > grid.arrange(p1, p2, p3, p4, nrow = 2, ncol = 2)
##
## > g <- arrangeGrob(p1, p2, p3, p4, nrow = 2, ncol = 2)
##
## > ggsave(file = "../output/figures/key_variables.png",
## + g)
## Saving 7 x 5 in image

source("../code/206_Confounder plots.R", echo = TRUE)
##
## > BRFSS <- read.csv(file = "../output/BRFSS.csv", header = TRUE,
## + sep = ",")
##
## > BRFSS$AGEG5YR2 <- as.factor(BRFSS$AGEG5YR2)
##
## > levels(BRFSS$AGEG5YR2) <- c("50-59", "60-69", "70-79",
## + "80+")
##
## > BRFSS$EDGRP <- as.factor(BRFSS$EDGRP)
##
## > levels(BRFSS$EDGRP) <- c("Grd 1-11", "HS Grad", "Some Coll",
## + "Coll Grad", "NR")
##
## > BRFSS$INCOME3 <- as.factor(BRFSS$INCOME3)
##
## > levels(BRFSS$INCOME3) <- c("<$15K", "$15to<$25", "$25to<$35K",
## + "$35to<$50K", "$50K+", "NR")
##
## > BRFSS$DENVST4 <- as.factor(BRFSS$DENVST4)
##
## > levels(BRFSS$DENVST4) <- c("Yes", "No", "NR")
##
## > p1 <- ggplot(BRFSS, aes(AGEG5YR2, fill = AGEG5YR2))
##
## > p1 <- p1 + geom_bar(color = "Black") + scale_fill_brewer(palette = "Blues") +
## + theme_minimal() + theme(axis.text.x = element_text(angle = 45)) .... [TRUNCATED]
##
## > p2 <- ggplot(BRFSS, aes(EDGRP, fill = EDGRP))
##
## > p2 <- p2 + geom_bar(color = "Black") + scale_fill_brewer(palette = "Blues") +
## + theme_minimal() + theme(axis.text.x = element_text(angle = 45)) .... [TRUNCATED]
##
## > p3 <- ggplot(BRFSS, aes(INCOME3, fill = INCOME3))
##
## > p3 <- p3 + geom_bar(color = "Black") + scale_fill_brewer(palette = "Blues") +
## + theme_minimal() + theme(axis.text.x = element_text(angle = 45)) .... [TRUNCATED]
##
## > p4 <- ggplot(BRFSS, aes(DENVST4, fill = DENVST4))
##
## > p4 <- p4 + geom_bar(color = "Black") + scale_fill_brewer(palette = "Blues") +
## + theme_minimal() + theme(axis.text.x = element_text(angle = 45)) .... [TRUNCATED]
##
## > grid.arrange(p1, p2, p3, p4, nrow = 2, ncol = 2)
##
## > g <- arrangeGrob(p1, p2, p3, p4, nrow = 2, ncol = 2)
##
## > ggsave(file = "../output/figures/confounders.png",
## + g)
## Saving 7 x 5 in image

source("../code/207_More confounder plots.R", echo = TRUE)
##
## > BRFSS <- read.csv(file = "../output/BRFSS.csv", header = TRUE,
## + sep = ",")
##
## > BRFSS$DIABETE4 <- as.factor(BRFSS$DIABETE4)
##
## > levels(BRFSS$DIABETE4) <- c("Yes", "No", "NR")
##
## > BRFSS$SMOKER4 <- as.factor(BRFSS$SMOKER4)
##
## > levels(BRFSS$SMOKER4) <- c("NvrSmk", "FrmSmk", "Smk",
## + "NR")
##
## > BRFSS$EXERANY3 <- as.factor(BRFSS$EXERANY3)
##
## > levels(BRFSS$EXERANY3) <- c("Yes", "No", "NR")
##
## > BRFSS$BMICAT <- as.factor(BRFSS$BMICAT)
##
## > levels(BRFSS$BMICAT) <- c("UndWt", "Norm", "OvrWt",
## + "Obese", "NR")
##
## > p1 <- ggplot(BRFSS, aes(DIABETE4, fill = DIABETE4))
##
## > p1 <- p1 + geom_bar(color = "Black") + scale_fill_brewer(palette = "Blues") +
## + theme_minimal() + theme(axis.text.x = element_text(angle = 45)) .... [TRUNCATED]
##
## > p2 <- ggplot(BRFSS, aes(SMOKER4, fill = SMOKER4))
##
## > p2 <- p2 + geom_bar(color = "Black") + scale_fill_brewer(palette = "Blues") +
## + theme_minimal() + theme(axis.text.x = element_text(angle = 45)) .... [TRUNCATED]
##
## > p3 <- ggplot(BRFSS, aes(EXERANY3, fill = EXERANY3))
##
## > p3 <- p3 + geom_bar(color = "Black") + scale_fill_brewer(palette = "Blues") +
## + theme_minimal() + theme(axis.text.x = element_text(angle = 45)) .... [TRUNCATED]
##
## > p4 <- ggplot(BRFSS, aes(BMICAT, fill = BMICAT))
##
## > p4 <- p4 + geom_bar(color = "Black") + scale_fill_brewer(palette = "Blues") +
## + theme_minimal() + theme(axis.text.x = element_text(angle = 45)) .... [TRUNCATED]
##
## > grid.arrange(p1, p2, p3, p4, nrow = 2, ncol = 2)
##
## > g <- arrangeGrob(p1, p2, p3, p4, nrow = 2, ncol = 2)
##
## > ggsave(file = "../output/figures/more_confounders.png",
## + g)
## Saving 7 x 5 in image

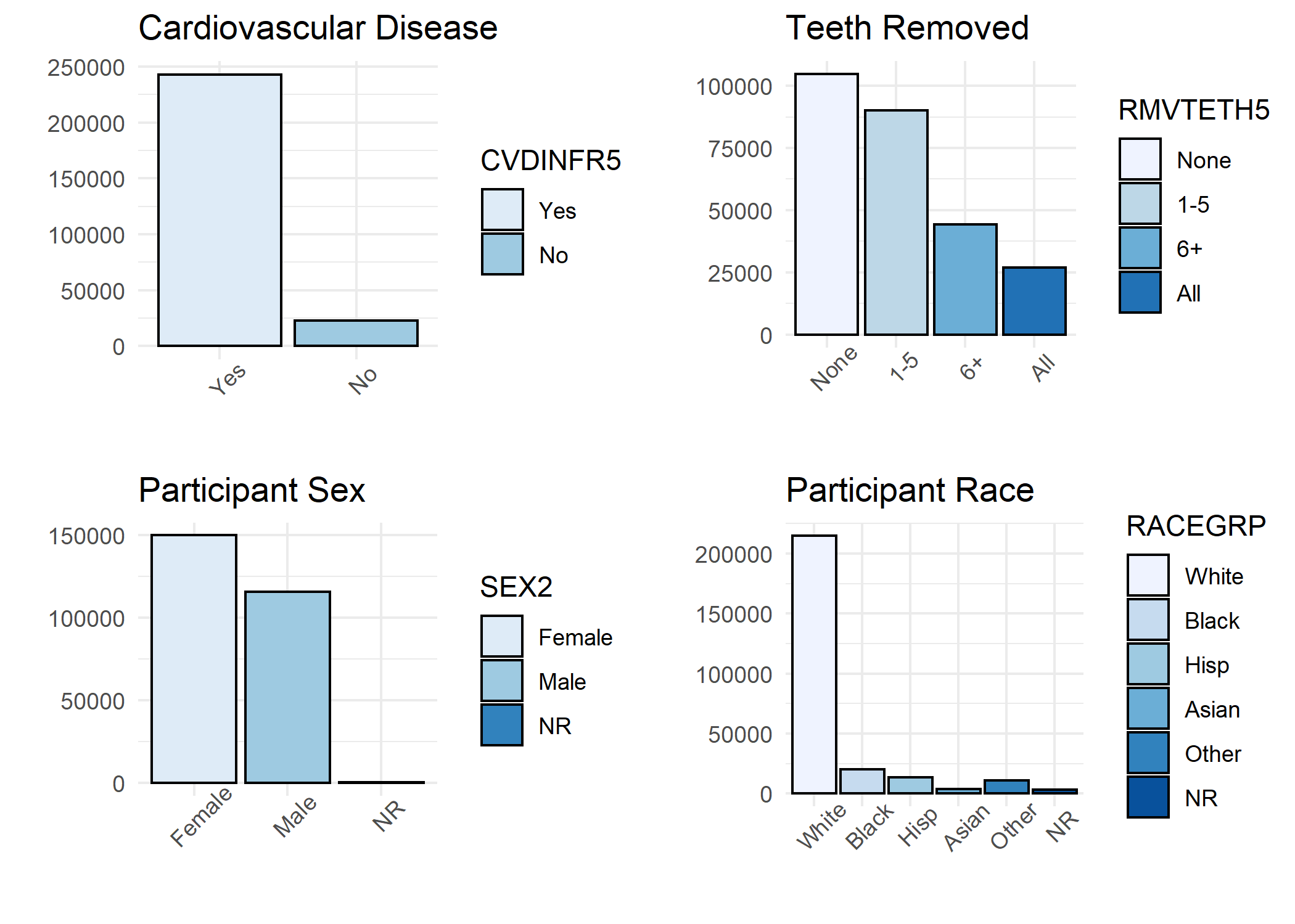
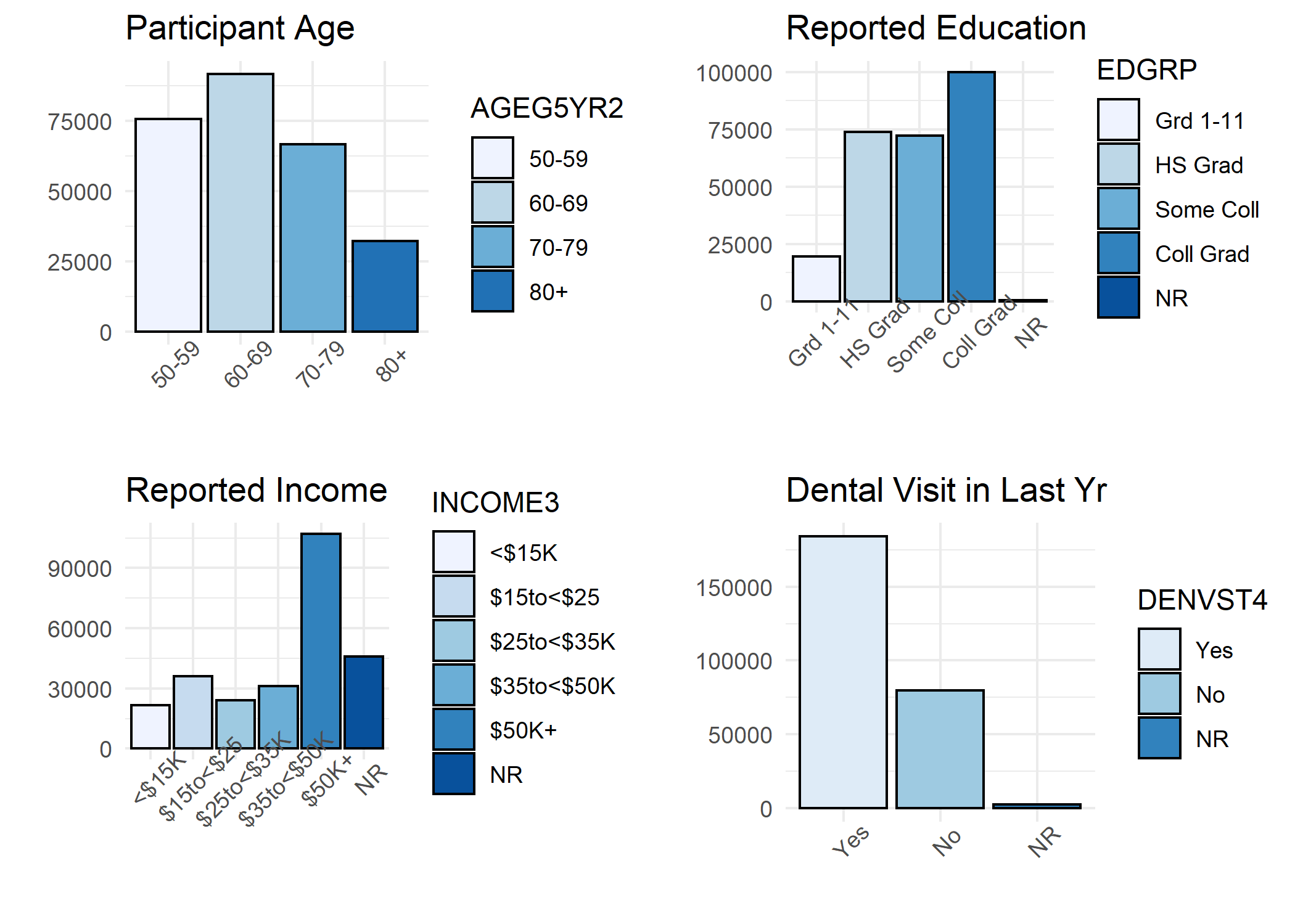
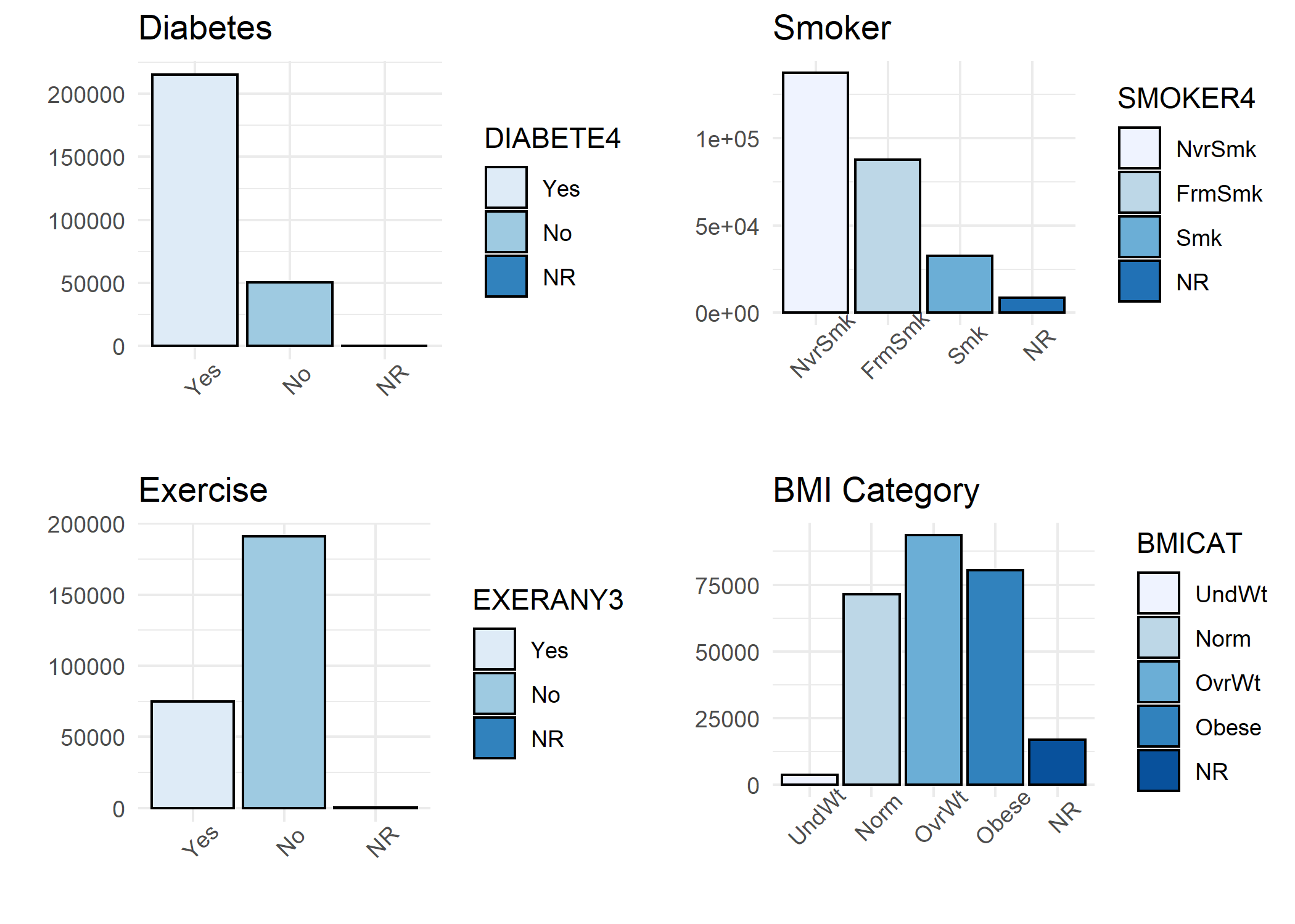
Step 3: Frequency Calculations for Table 1
Next, the frequencies were calculated for each variable. These result appear in table 1 below.
3 sets of variable values were examined:
- Total frequencies for each variable
- Frequencies each variable for those reporting Cardiovascular Disease Only
- Frequencies each variable for those reporting No Cardiovascular Disease Only
source("../code/210_Table 1 overall frequencies.R", echo = TRUE)
##
## > BRFSS <- read.csv(file = "../output/BRFSS.csv", header = TRUE,
## + sep = ",")
##
## > cvdFreq <- table(BRFSS$CVDINFR5)
##
## > cvdFreq
##
## 0 1
## 243070 23039
##
## > write.csv(cvdFreq, file = "../output/frequencies/overall/cvdFreq.csv")
##
## > FreqTbl <- defmacro(OutputTable, InputVar, CSVTable,
## + expr = {
## + OutputTable <- table(InputVar)
## + write.csv(OutputTable, file .... [TRUNCATED]
##
## > FreqTbl(teethFreq, BRFSS$RMVTETH5, "teeth")
##
## > FreqTbl(sexFreq, BRFSS$SEX2, "sex")
##
## > FreqTbl(hispFreq, BRFSS$HISPANIC, "hispanic")
##
## > FreqTbl(raceFreq, BRFSS$RACEGRP, "race")
##
## > FreqTbl(sexFreq, BRFSS$AGEG5YR2, "age")
##
## > FreqTbl(eduFreq, BRFSS$EDGRP, "edu")
##
## > FreqTbl(incFreq, BRFSS$INCOME3, "inc")
##
## > FreqTbl(denvstFreq, BRFSS$DENVST4, "denvst")
##
## > FreqTbl(diabFreq, BRFSS$DIABETE4, "diab")
##
## > FreqTbl(smokeFreq, BRFSS$SMOKER4, "smoke")
##
## > FreqTbl(exFreq, BRFSS$EXERANY3, "ex")
##
## > FreqTbl(bmiFreq, BRFSS$BMICAT, "bmi")
source("../code/215_Table 1 cv frequencies.R", echo = TRUE)
##
## > BRFSS <- read.csv(file = "../output/BRFSS.csv", header = TRUE,
## + sep = ",")
##
## > cv_only <- subset(BRFSS, CVDINFR5 == 0)
##
## > table(cv_only$RMVTETH5)
##
## 0 1 2 3
## 99474 83224 38416 21956
##
## > nrow(cv_only)
## [1] 243070
##
## > FreqTbl <- defmacro(OutputTable, InputVar, CSVTable,
## + expr = {
## + OutputTable <- table(InputVar)
## + write.csv(OutputTable, file .... [TRUNCATED]
##
## > FreqTbl(teethFreq, cv_only$RMVTETH5, "teeth")
##
## > FreqTbl(sexFreq, cv_only$SEX2, "sex")
##
## > FreqTbl(hispFreq, cv_only$HISPANIC, "hispanic")
##
## > FreqTbl(raceFreq, cv_only$RACEGRP, "race")
##
## > FreqTbl(sexFreq, cv_only$AGEG5YR2, "age")
##
## > FreqTbl(eduFreq, cv_only$EDGRP, "edu")
##
## > FreqTbl(incFreq, cv_only$INCOME3, "inc")
##
## > FreqTbl(denvstFreq, cv_only$DENVST4, "denvst")
##
## > FreqTbl(diabFreq, cv_only$DIABETE4, "diab")
##
## > FreqTbl(smokeFreq, cv_only$SMOKER4, "smoke")
##
## > FreqTbl(exFreq, cv_only$EXERANY3, "ex")
##
## > FreqTbl(bmiFreq, cv_only$BMICAT, "bmi")
source("../code/220_Table 1 nocv frequencies.R", echo = TRUE)
##
## > BRFSS <- read.csv(file = "../output/BRFSS.csv", header = TRUE,
## + sep = ",")
##
## > nocv_only <- subset(BRFSS, CVDINFR5 == 1)
##
## > table(nocv_only$RMVTETH5)
##
## 0 1 2 3
## 5328 6968 5901 4842
##
## > nrow(nocv_only)
## [1] 23039
##
## > FreqTbl <- defmacro(OutputTable, InputVar, CSVTable,
## + expr = {
## + OutputTable <- table(InputVar)
## + write.csv(OutputTable, file .... [TRUNCATED]
##
## > FreqTbl(teethFreq, nocv_only$RMVTETH5, "teeth")
##
## > FreqTbl(sexFreq, nocv_only$SEX2, "sex")
##
## > FreqTbl(hispFreq, nocv_only$HISPANIC, "hispanic")
##
## > FreqTbl(raceFreq, nocv_only$RACEGRP, "race")
##
## > FreqTbl(ageFreq, nocv_only$AGEG5YR2, "age")
##
## > FreqTbl(eduFreq, nocv_only$EDGRP, "edu")
##
## > FreqTbl(incFreq, nocv_only$INCOME3, "inc")
##
## > FreqTbl(denvstFreq, nocv_only$DENVST4, "denvst")
##
## > FreqTbl(diabFreq, nocv_only$DIABETE4, "diab")
##
## > FreqTbl(smokeFreq, nocv_only$SMOKER4, "smoke")
##
## > FreqTbl(exFreq, nocv_only$EXERANY3, "ex")
##
## > FreqTbl(bmiFreq, nocv_only$BMICAT, "bmi")
Step 4: Calculation of Chi-Squares
Chi-square values were calcualted for all variables. All were significant.
source("../code/230_Table 1 chi squares.R", echo = TRUE)
##
## > BRFSS <- read.csv(file = "../data/BRFSS.csv", header = TRUE,
## + sep = ",")
##
## > teethTbl = table(BRFSS$CVDINFR5, BRFSS$RMVTETH5)
##
## > chisq.test(teethTbl)
##
## Pearson's Chi-squared test
##
## data: teethTbl
## X-squared = 6008.5, df = 3, p-value < 2.2e-16
##
##
## > ChiTest <- defmacro(VarName, TblName, expr = {
## + TblName = table(BRFSS$CVDINFR5, BRFSS$VarName)
## + chisq.test(TblName)
## + })
##
## > ChiTest(RMVTETH5, teethTbl)
##
## Pearson's Chi-squared test
##
## data: teethTbl
## X-squared = 6008.5, df = 3, p-value < 2.2e-16
##
##
## > ChiTest(SEX2, sexTbl)
##
## Pearson's Chi-squared test
##
## data: sexTbl
## X-squared = 2852.8, df = 2, p-value < 2.2e-16
##
##
## > ChiTest(HISPANIC, hispanicTbl)
##
## Pearson's Chi-squared test
##
## data: hispanicTbl
## X-squared = 33.897, df = 2, p-value = 4.358e-08
##
##
## > ChiTest(RACEGRP, raceTbl)
##
## Pearson's Chi-squared test
##
## data: raceTbl
## X-squared = 330.57, df = 5, p-value < 2.2e-16
##
##
## > ChiTest(AGEG5YR2, ageTbl)
##
## Pearson's Chi-squared test
##
## data: ageTbl
## X-squared = 3482.2, df = 3, p-value < 2.2e-16
##
##
## > ChiTest(EDGRP, eduTbl)
##
## Pearson's Chi-squared test
##
## data: eduTbl
## X-squared = 2193.5, df = 4, p-value < 2.2e-16
##
##
## > ChiTest(INCOME3, EdTbl)
##
## Pearson's Chi-squared test
##
## data: EdTbl
## X-squared = 3066.2, df = 5, p-value < 2.2e-16
##
##
## > ChiTest(DENVST4, denvstTbl)
##
## Pearson's Chi-squared test
##
## data: denvstTbl
## X-squared = 2535.8, df = 2, p-value < 2.2e-16
##
##
## > ChiTest(DIABETE4, diabTbl)
##
## Pearson's Chi-squared test
##
## data: diabTbl
## X-squared = 4665.9, df = 2, p-value < 2.2e-16
##
##
## > ChiTest(SMOKER4, smokeTbl)
##
## Pearson's Chi-squared test
##
## data: smokeTbl
## X-squared = 2661.8, df = 3, p-value < 2.2e-16
##
##
## > ChiTest(EXERANY3, exerTbl)
##
## Pearson's Chi-squared test
##
## data: exerTbl
## X-squared = 1668, df = 2, p-value < 2.2e-16
##
##
## > ChiTest(BMICAT, bmiTbl)
##
## Pearson's Chi-squared test
##
## data: bmiTbl
## X-squared = 640.78, df = 4, p-value < 2.2e-16
Step 5: Create Final Table 1
The table below shows the participant counts (frequencies) from the descriptive analysis arrange in a complet Table 1. The table also shows relative percentages, and chi-square significance values. All variables were significant at the p<0.001 level.
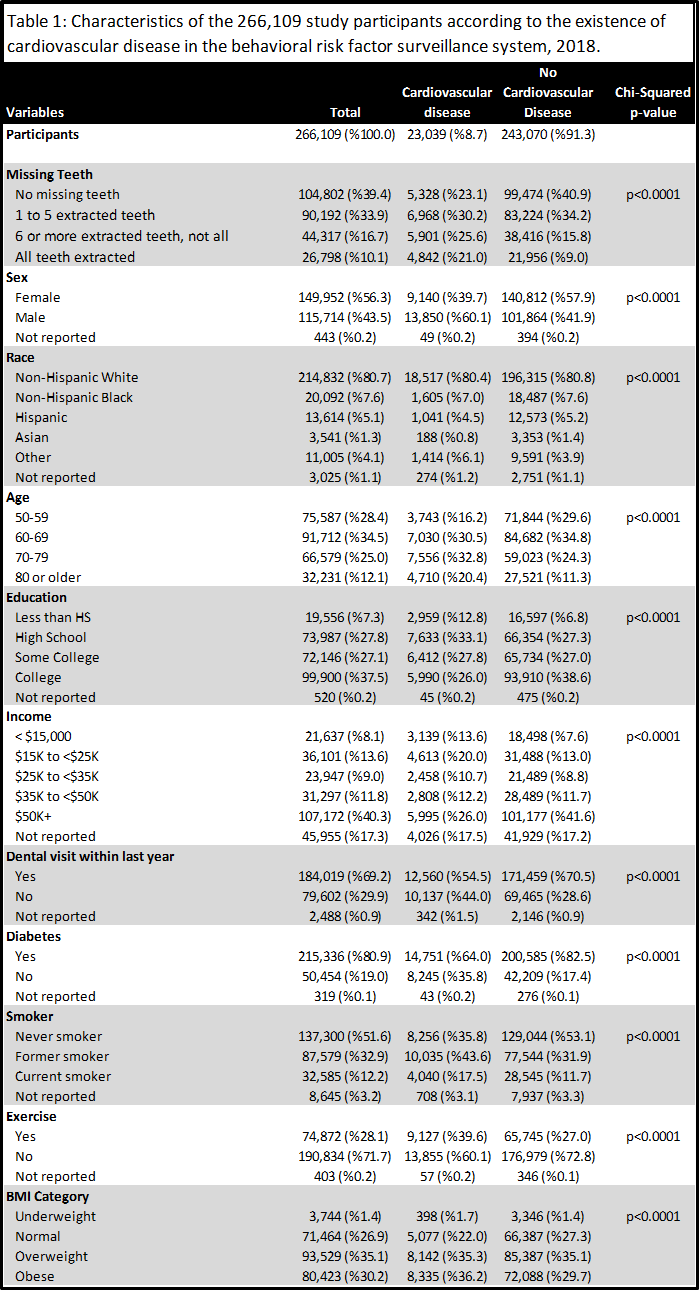
This concluded the descriptive analysis.
Return to the Main Overview