Linear Regression
First created on Nov 8, 2019. Updated on Nov 11, 2019
Step 1: Load Libraries
source("../code/100_Dependencies.R", echo = TRUE)
##
## > library(foreign)
##
## > library(Hmisc)
## Loading required package: lattice
## Loading required package: survival
## Loading required package: Formula
## Loading required package: ggplot2
##
## Attaching package: 'Hmisc'
## The following objects are masked from 'package:base':
##
## format.pval, units
##
## > library(e1071)
##
## Attaching package: 'e1071'
## The following object is masked from 'package:Hmisc':
##
## impute
##
## > library(gtools)
##
## Attaching package: 'gtools'
## The following object is masked from 'package:e1071':
##
## permutations
##
## > library(MASS)
##
## > library(ggplot2)
##
## > library(gridExtra)
##
## > library(broom)
##
## > library(scales)
##
## > library(ggfortify)
##
## > library(gvlma)
Step 2: Run Linear Regression Models
source("../code/260_Linear regression models.R", echo = TRUE)
##
## > BRFSS <- read.csv(file = "../output/BRFSS.csv", header = TRUE,
## + sep = ",")
##
## > str(BRFSS)
## 'data.frame': 266109 obs. of 52 variables:
## $ CVDINFR4 : int 2 2 2 2 2 2 2 2 2 2 ...
## $ RMVTETH4 : int 1 1 8 8 2 8 1 2 1 3 ...
## $ SEX1 : int 2 1 2 1 1 2 2 2 2 1 ...
## $ X_HISPANC: int 2 2 2 2 2 2 2 2 2 2 ...
## $ X_MRACE1 : int 1 1 1 1 1 1 1 2 1 1 ...
## $ X_AGEG5YR: int 13 10 12 8 13 8 11 12 10 11 ...
## $ EDUCA : int 6 4 6 4 4 5 4 3 3 5 ...
## $ INCOME2 : int 6 3 8 8 4 6 4 2 2 7 ...
## $ X_DENVST3: int 1 2 1 1 1 1 2 1 1 2 ...
## $ DIABETE3 : int 3 3 3 1 3 3 1 3 1 3 ...
## $ X_SMOKER3: int 4 4 4 3 3 4 4 4 4 4 ...
## $ EXERANY2 : int 2 1 1 2 2 1 2 2 1 1 ...
## $ X_BMI5CAT: int 2 3 2 3 3 4 4 4 4 4 ...
## $ CVDINFR5 : int 0 0 0 0 0 0 0 0 0 0 ...
## $ RMVTETH5 : int 1 1 0 0 2 0 1 2 1 3 ...
## $ MOSTTEETH: int 1 1 0 0 0 0 1 0 1 0 ...
## $ FEWTEETH : int 0 0 0 0 1 0 0 1 0 0 ...
## $ NOTEETH : int 0 0 0 0 0 0 0 0 0 1 ...
## $ SEX2 : int 0 1 0 1 1 0 0 0 0 1 ...
## $ MALE : int 0 1 0 1 1 0 0 0 0 1 ...
## $ HISPANC2 : int 0 0 0 0 0 0 0 0 0 0 ...
## $ HISPANIC : int 0 0 0 0 0 0 0 0 0 0 ...
## $ RACEGRP : int 0 0 0 0 0 0 0 1 0 0 ...
## $ BLACK : int 0 0 0 0 0 0 0 1 0 0 ...
## $ ASIAN : int 0 0 0 0 0 0 0 0 0 0 ...
## $ OTHRACE : int 0 0 0 0 0 0 0 0 0 0 ...
## $ AGEG5YR2 : int 3 1 2 0 3 0 2 2 1 2 ...
## $ AGE1 : int 0 1 0 0 0 0 0 0 1 0 ...
## $ AGE2 : int 0 0 1 0 0 0 1 1 0 1 ...
## $ AGE3 : int 1 0 0 0 1 0 0 0 0 0 ...
## $ EDGRP : int 3 1 3 1 1 2 1 0 0 2 ...
## $ LOWED : int 0 0 0 0 0 0 0 1 1 0 ...
## $ SOMECOLL : int 0 0 0 0 0 1 0 0 0 1 ...
## $ COLLEGE : int 1 0 1 0 0 0 0 0 0 0 ...
## $ INCOME3 : int 3 1 4 4 1 3 1 0 0 4 ...
## $ INC1 : int 0 0 0 0 0 0 0 1 1 0 ...
## $ INC2 : int 0 1 0 0 1 0 1 0 0 0 ...
## $ INC3 : int 0 0 0 0 0 0 0 0 0 0 ...
## $ INC4 : int 1 0 0 0 0 1 0 0 0 0 ...
## $ DENVST4 : int 0 1 0 0 0 0 1 0 0 1 ...
## $ NODENVST : int 0 1 0 0 0 0 1 0 0 1 ...
## $ DIABETE4 : int 0 0 0 1 0 0 1 0 1 0 ...
## $ NODIABETE: int 1 1 1 0 1 1 0 1 0 1 ...
## $ SMOKER4 : int 0 0 0 1 1 0 0 0 0 0 ...
## $ FRMRSMK : int 0 0 0 1 1 0 0 0 0 0 ...
## $ SMOKE : int 0 0 0 0 0 0 0 0 0 0 ...
## $ EXERANY3 : int 0 1 1 0 0 1 0 0 1 1 ...
## $ NOEXER : int 1 0 0 1 1 0 1 1 0 0 ...
## $ BMICAT : int 1 2 1 2 2 3 3 3 3 3 ...
## $ UNDWT : int 0 0 0 0 0 0 0 0 0 0 ...
## $ OVWT : int 0 1 0 1 1 0 0 0 0 0 ...
## $ OBESE : int 0 0 0 0 0 1 1 1 1 1 ...
##
## > summary(BRFSS)
## CVDINFR4 RMVTETH4 SEX1 X_HISPANC
## Min. :1.000 Min. :1.000 Min. :1.000 Min. :1.000
## 1st Qu.:2.000 1st Qu.:1.000 1st Qu.:1.000 1st Qu.:2.000
## Median :2.000 Median :2.000 Median :2.000 Median :2.000
## Mean :1.913 Mean :4.125 Mean :1.575 Mean :2.005
## 3rd Qu.:2.000 3rd Qu.:8.000 3rd Qu.:2.000 3rd Qu.:2.000
## Max. :2.000 Max. :8.000 Max. :9.000 Max. :9.000
##
## X_MRACE1 X_AGEG5YR EDUCA INCOME2
## Min. : 1.00 Min. : 7.000 Min. :1.000 Min. : 1.00
## 1st Qu.: 1.00 1st Qu.: 8.000 1st Qu.:4.000 1st Qu.: 5.00
## Median : 1.00 Median :10.000 Median :5.000 Median : 7.00
## Mean : 2.83 Mean : 9.847 Mean :4.931 Mean :19.87
## 3rd Qu.: 1.00 3rd Qu.:11.000 3rd Qu.:6.000 3rd Qu.: 8.00
## Max. :99.00 Max. :13.000 Max. :9.000 Max. :99.00
## NA's :2 NA's :3 NA's :1983
## X_DENVST3 DIABETE3 X_SMOKER3 EXERANY2
## Min. :1.000 Min. :1.000 Min. :1.000 Min. :1.000
## 1st Qu.:1.000 1st Qu.:3.000 1st Qu.:3.000 1st Qu.:1.000
## Median :1.000 Median :3.000 Median :4.000 Median :1.000
## Mean :1.374 Mean :2.643 Mean :3.498 Mean :1.291
## 3rd Qu.:2.000 3rd Qu.:3.000 3rd Qu.:4.000 3rd Qu.:2.000
## Max. :9.000 Max. :9.000 Max. :9.000 Max. :9.000
## NA's :2
## X_BMI5CAT CVDINFR5 RMVTETH5 MOSTTEETH
## Min. :1.000 Min. :0.00000 Min. :0.0000 Min. :0.0000
## 1st Qu.:2.000 1st Qu.:0.00000 1st Qu.:0.0000 1st Qu.:0.0000
## Median :3.000 Median :0.00000 Median :1.0000 Median :0.0000
## Mean :3.006 Mean :0.08658 Mean :0.9741 Mean :0.3389
## 3rd Qu.:4.000 3rd Qu.:0.00000 3rd Qu.:2.0000 3rd Qu.:1.0000
## Max. :4.000 Max. :1.00000 Max. :3.0000 Max. :1.0000
## NA's :16949
## FEWTEETH NOTEETH SEX2 MALE
## Min. :0.0000 Min. :0.0000 Min. :0.0000 Min. :0.0000
## 1st Qu.:0.0000 1st Qu.:0.0000 1st Qu.:0.0000 1st Qu.:0.0000
## Median :0.0000 Median :0.0000 Median :0.0000 Median :0.0000
## Mean :0.1665 Mean :0.1007 Mean :0.4498 Mean :0.4348
## 3rd Qu.:0.0000 3rd Qu.:0.0000 3rd Qu.:1.0000 3rd Qu.:1.0000
## Max. :1.0000 Max. :1.0000 Max. :9.0000 Max. :1.0000
##
## HISPANC2 HISPANIC RACEGRP BLACK
## Min. :0.0000 Min. :0.00000 Min. :0.0000 Min. :0.0000
## 1st Qu.:0.0000 1st Qu.:0.00000 1st Qu.:0.0000 1st Qu.:0.0000
## Median :0.0000 Median :0.00000 Median :0.0000 Median :0.0000
## Mean :0.1231 Mean :0.05116 Mean :0.4855 Mean :0.0755
## 3rd Qu.:0.0000 3rd Qu.:0.00000 3rd Qu.:0.0000 3rd Qu.:0.0000
## Max. :9.0000 Max. :1.00000 Max. :9.0000 Max. :1.0000
##
## ASIAN OTHRACE AGEG5YR2 AGE1
## Min. :0.00000 Min. :0.00000 Min. :0.000 Min. :0.0000
## 1st Qu.:0.00000 1st Qu.:0.00000 1st Qu.:0.000 1st Qu.:0.0000
## Median :0.00000 Median :0.00000 Median :1.000 Median :0.0000
## Mean :0.01331 Mean :0.04136 Mean :1.208 Mean :0.3446
## 3rd Qu.:0.00000 3rd Qu.:0.00000 3rd Qu.:2.000 3rd Qu.:1.0000
## Max. :1.00000 Max. :1.00000 Max. :3.000 Max. :1.0000
##
## AGE2 AGE3 EDGRP LOWED
## Min. :0.0000 Min. :0.0000 Min. :0.000 Min. :0.00000
## 1st Qu.:0.0000 1st Qu.:0.0000 1st Qu.:1.000 1st Qu.:0.00000
## Median :0.0000 Median :0.0000 Median :2.000 Median :0.00000
## Mean :0.2502 Mean :0.1211 Mean :1.964 Mean :0.07349
## 3rd Qu.:1.0000 3rd Qu.:0.0000 3rd Qu.:3.000 3rd Qu.:0.00000
## Max. :1.0000 Max. :1.0000 Max. :9.000 Max. :1.00000
##
## SOMECOLL COLLEGE INCOME3 INC1
## Min. :0.0000 Min. :0.0000 Min. :0.000 Min. :0.00000
## 1st Qu.:0.0000 1st Qu.:0.0000 1st Qu.:2.000 1st Qu.:0.00000
## Median :0.0000 Median :0.0000 Median :4.000 Median :0.00000
## Mean :0.2711 Mean :0.3754 Mean :3.834 Mean :0.08131
## 3rd Qu.:1.0000 3rd Qu.:1.0000 3rd Qu.:4.000 3rd Qu.:0.00000
## Max. :1.0000 Max. :1.0000 Max. :9.000 Max. :1.00000
##
## INC2 INC3 INC4 DENVST4
## Min. :0.0000 Min. :0.00000 Min. :0.0000 Min. :0.0000
## 1st Qu.:0.0000 1st Qu.:0.00000 1st Qu.:0.0000 1st Qu.:0.0000
## Median :0.0000 Median :0.00000 Median :0.0000 Median :0.0000
## Mean :0.1357 Mean :0.08999 Mean :0.1176 Mean :0.3833
## 3rd Qu.:0.0000 3rd Qu.:0.00000 3rd Qu.:0.0000 3rd Qu.:1.0000
## Max. :1.0000 Max. :1.00000 Max. :1.0000 Max. :9.0000
##
## NODENVST DIABETE4 NODIABETE SMOKER4
## Min. :0.0000 Min. :0.0000 Min. :0.0000 Min. :0.0000
## 1st Qu.:0.0000 1st Qu.:0.0000 1st Qu.:1.0000 1st Qu.:0.0000
## Median :0.0000 Median :0.0000 Median :1.0000 Median :0.0000
## Mean :0.2991 Mean :0.2004 Mean :0.8092 Mean :0.8664
## 3rd Qu.:1.0000 3rd Qu.:0.0000 3rd Qu.:1.0000 3rd Qu.:1.0000
## Max. :1.0000 Max. :9.0000 Max. :1.0000 Max. :9.0000
##
## FRMRSMK SMOKE EXERANY3 NOEXER
## Min. :0.0000 Min. :0.0000 Min. :0.0000 Min. :0.0000
## 1st Qu.:0.0000 1st Qu.:0.0000 1st Qu.:0.0000 1st Qu.:0.0000
## Median :0.0000 Median :0.0000 Median :1.0000 Median :0.0000
## Mean :0.3291 Mean :0.1224 Mean :0.7308 Mean :0.2814
## 3rd Qu.:1.0000 3rd Qu.:0.0000 3rd Qu.:1.0000 3rd Qu.:1.0000
## Max. :1.0000 Max. :1.0000 Max. :9.0000 Max. :1.0000
##
## BMICAT UNDWT OVWT OBESE
## Min. :0.000 Min. :0.00000 Min. :0.0000 Min. :0.0000
## 1st Qu.:1.000 1st Qu.:0.00000 1st Qu.:0.0000 1st Qu.:0.0000
## Median :2.000 Median :0.00000 Median :0.0000 Median :0.0000
## Mean :2.451 Mean :0.01407 Mean :0.3515 Mean :0.3022
## 3rd Qu.:3.000 3rd Qu.:0.00000 3rd Qu.:1.0000 3rd Qu.:1.0000
## Max. :9.000 Max. :1.00000 Max. :1.0000 Max. :1.0000
##
##
## > model1 <- lm(CVDINFR5 ~ NOTEETH + FEWTEETH + MOSTTEETH,
## + data = BRFSS)
##
## > summary(model1)
##
## Call:
## lm(formula = CVDINFR5 ~ NOTEETH + FEWTEETH + MOSTTEETH, data = BRFSS)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.18069 -0.07726 -0.07726 -0.05084 0.94916
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.0508387 0.0008588 59.20 <2e-16 ***
## NOTEETH 0.1298464 0.0019032 68.23 <2e-16 ***
## FEWTEETH 0.0823156 0.0015754 52.25 <2e-16 ***
## MOSTTEETH 0.0264187 0.0012628 20.92 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.278 on 266105 degrees of freedom
## Multiple R-squared: 0.02258, Adjusted R-squared: 0.02257
## F-statistic: 2049 on 3 and 266105 DF, p-value: < 2.2e-16
##
##
## > model2 <- lm(CVDINFR5 ~ NOTEETH + FEWTEETH + MOSTTEETH +
## + MALE + AGE1 + AGE2 + AGE3, data = BRFSS)
##
## > summary(model2)
##
## Call:
## lm(formula = CVDINFR5 ~ NOTEETH + FEWTEETH + MOSTTEETH + MALE +
## AGE1 + AGE2 + AGE3, data = BRFSS)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.25791 -0.11350 -0.07078 -0.02102 0.99960
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.0004043 0.0012393 0.326 0.744
## NOTEETH 0.1148229 0.0019111 60.083 <2e-16 ***
## FEWTEETH 0.0695639 0.0015779 44.086 <2e-16 ***
## MOSTTEETH 0.0189619 0.0012570 15.084 <2e-16 ***
## MALE 0.0616769 0.0010787 57.177 <2e-16 ***
## AGE1 0.0206114 0.0013573 15.185 <2e-16 ***
## AGE2 0.0514180 0.0014826 34.682 <2e-16 ***
## AGE3 0.0810020 0.0018611 43.523 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.2753 on 266101 degrees of freedom
## Multiple R-squared: 0.04193, Adjusted R-squared: 0.04191
## F-statistic: 1664 on 7 and 266101 DF, p-value: < 2.2e-16
##
##
## > min.model <- lm(CVDINFR5 ~ 1, data = BRFSS)
##
## > fwd.model <- step(min.model, direction = "forward",
## + scope = (~NOTEETH + FEWTEETH + MOSTTEETH + MALE + BLACK +
## + HISPANIC + ASIAN + O .... [TRUNCATED]
## Start: AIC=-675189.5
## CVDINFR5 ~ 1
##
## Df Sum of Sq RSS AIC
## + NODIABETE 1 368.73 20676 -679891
## + NOTEETH 1 263.91 20780 -678546
## + MALE 1 224.51 20820 -678042
## + NODENVST 1 188.77 20856 -677585
## + AGE3 1 130.07 20914 -676837
## + NOEXER 1 130.00 20914 -676836
## + FEWTEETH 1 115.35 20929 -676650
## + COLLEGE 1 113.32 20931 -676624
## + FRMRSMK 1 102.38 20942 -676485
## + LOWED 1 88.44 20956 -676308
## + INC1 1 80.60 20964 -676209
## + INC2 1 70.91 20973 -676086
## + AGE2 1 64.31 20980 -676002
## + SMOKE 1 51.96 20992 -675845
## + OBESE 1 33.55 21011 -675612
## + OTHRACE 1 20.16 21024 -675443
## + AGE1 1 13.78 21031 -675362
## + MOSTTEETH 1 11.85 21033 -675337
## + INC3 1 6.79 21038 -675273
## + ASIAN 1 4.02 21040 -675238
## + UNDWT 1 1.48 21043 -675206
## + HISPANIC 1 1.47 21043 -675206
## + BLACK 1 0.97 21043 -675200
## + SOMECOLL 1 0.52 21044 -675194
## + INC4 1 0.35 21044 -675192
## <none> 21044 -675189
## + OVWT 1 0.03 21044 -675188
##
## Step: AIC=-679891.4
## CVDINFR5 ~ NODIABETE
##
## Df Sum of Sq RSS AIC
## + NOTEETH 1 211.087 20465 -682620
## + MALE 1 202.937 20473 -682514
## + NODENVST 1 143.687 20532 -681745
## + AGE3 1 129.094 20547 -681556
## + FRMRSMK 1 88.281 20587 -681028
## + FEWTEETH 1 82.884 20593 -680958
## + NOEXER 1 80.517 20595 -680928
## + COLLEGE 1 76.909 20599 -680881
## + LOWED 1 62.071 20614 -680689
## + INC1 1 56.058 20620 -680612
## + SMOKE 1 52.629 20623 -680568
## + INC2 1 49.904 20626 -680532
## + AGE2 1 46.569 20629 -680489
## + AGE1 1 15.611 20660 -680090
## + OTHRACE 1 14.025 20662 -680070
## + MOSTTEETH 1 11.735 20664 -680040
## + BLACK 1 6.488 20669 -679973
## + ASIAN 1 4.985 20671 -679954
## + INC3 1 4.820 20671 -679951
## + HISPANIC 1 4.463 20671 -679947
## + OBESE 1 4.070 20672 -679942
## + UNDWT 1 3.665 20672 -679937
## + OVWT 1 1.307 20674 -679906
## + INC4 1 0.288 20675 -679893
## + SOMECOLL 1 0.170 20676 -679892
## <none> 20676 -679891
##
## Step: AIC=-682620.2
## CVDINFR5 ~ NODIABETE + NOTEETH
##
## Df Sum of Sq RSS AIC
## + MALE 1 210.625 20254 -685371
## + FEWTEETH 1 133.633 20331 -684362
## + AGE3 1 102.134 20362 -683950
## + FRMRSMK 1 74.213 20390 -683585
## + NODENVST 1 62.821 20402 -683436
## + NOEXER 1 51.750 20413 -683292
## + COLLEGE 1 39.824 20425 -683137
## + AGE2 1 36.410 20428 -683092
## + INC1 1 31.284 20433 -683025
## + LOWED 1 30.382 20434 -683014
## + INC2 1 29.800 20435 -683006
## + SMOKE 1 25.966 20439 -682956
## + AGE1 1 12.265 20452 -682778
## + OTHRACE 1 11.073 20454 -682762
## + BLACK 1 8.795 20456 -682733
## + OBESE 1 4.236 20460 -682673
## + ASIAN 1 3.460 20461 -682663
## + INC3 1 3.274 20461 -682661
## + HISPANIC 1 3.002 20462 -682657
## + OVWT 1 1.892 20463 -682643
## + UNDWT 1 1.854 20463 -682642
## + INC4 1 0.677 20464 -682627
## + SOMECOLL 1 0.475 20464 -682624
## <none> 20465 -682620
## + MOSTTEETH 1 0.005 20465 -682618
##
## Step: AIC=-685371.2
## CVDINFR5 ~ NODIABETE + NOTEETH + MALE
##
## Df Sum of Sq RSS AIC
## + FEWTEETH 1 132.644 20121 -687118
## + AGE3 1 116.751 20137 -686908
## + NOEXER 1 63.983 20190 -686211
## + NODENVST 1 52.828 20201 -686064
## + FRMRSMK 1 51.658 20202 -686049
## + COLLEGE 1 49.016 20205 -686014
## + AGE2 1 40.027 20214 -685896
## + INC1 1 37.418 20217 -685861
## + INC2 1 36.595 20217 -685850
## + LOWED 1 28.902 20225 -685749
## + SMOKE 1 21.759 20232 -685655
## + AGE1 1 14.691 20239 -685562
## + OTHRACE 1 9.835 20244 -685498
## + BLACK 1 6.152 20248 -685450
## + ASIAN 1 4.270 20250 -685425
## + INC3 1 4.071 20250 -685423
## + UNDWT 1 4.017 20250 -685422
## + HISPANIC 1 3.150 20251 -685411
## + OBESE 1 2.702 20251 -685405
## + SOMECOLL 1 1.685 20252 -685391
## + INC4 1 0.517 20253 -685376
## + OVWT 1 0.306 20254 -685373
## <none> 20254 -685371
## + MOSTTEETH 1 0.039 20254 -685370
##
## Step: AIC=-687117.7
## CVDINFR5 ~ NODIABETE + NOTEETH + MALE + FEWTEETH
##
## Df Sum of Sq RSS AIC
## + AGE3 1 99.053 20022 -688429
## + NOEXER 1 44.694 20077 -687707
## + FRMRSMK 1 40.261 20081 -687649
## + AGE2 1 32.074 20089 -687540
## + NODENVST 1 31.742 20090 -687536
## + COLLEGE 1 25.519 20096 -687453
## + INC1 1 23.471 20098 -687426
## + INC2 1 23.157 20098 -687422
## + MOSTTEETH 1 19.789 20102 -687378
## + LOWED 1 17.780 20104 -687351
## + AGE1 1 12.870 20108 -687286
## + BLACK 1 11.415 20110 -687267
## + SMOKE 1 10.357 20111 -687253
## + OTHRACE 1 7.686 20114 -687217
## + ASIAN 1 3.561 20118 -687163
## + UNDWT 1 3.313 20118 -687160
## + HISPANIC 1 3.150 20118 -687157
## + INC3 1 2.188 20119 -687145
## + OBESE 1 1.617 20120 -687137
## + SOMECOLL 1 1.232 20120 -687132
## + INC4 1 0.347 20121 -687120
## + OVWT 1 0.210 20121 -687118
## <none> 20121 -687118
##
## Step: AIC=-688429
## CVDINFR5 ~ NODIABETE + NOTEETH + MALE + FEWTEETH + AGE3
##
## Df Sum of Sq RSS AIC
## + AGE2 1 66.358 19956 -689310
## + NOEXER 1 37.409 19985 -688925
## + FRMRSMK 1 35.856 19986 -688904
## + NODENVST 1 34.865 19987 -688891
## + INC1 1 26.177 19996 -688775
## + COLLEGE 1 23.548 19999 -688740
## + SMOKE 1 20.515 20002 -688700
## + INC2 1 19.655 20003 -688688
## + LOWED 1 16.173 20006 -688642
## + MOSTTEETH 1 15.887 20006 -688638
## + OTHRACE 1 9.429 20013 -688552
## + BLACK 1 8.856 20013 -688545
## + OBESE 1 5.542 20017 -688501
## + ASIAN 1 3.267 20019 -688470
## + UNDWT 1 2.181 20020 -688456
## + HISPANIC 1 1.845 20020 -688451
## + SOMECOLL 1 1.750 20021 -688450
## + INC3 1 1.191 20021 -688443
## + AGE1 1 0.948 20021 -688440
## + OVWT 1 0.393 20022 -688432
## + INC4 1 0.247 20022 -688430
## <none> 20022 -688429
##
## Step: AIC=-689310.4
## CVDINFR5 ~ NODIABETE + NOTEETH + MALE + FEWTEETH + AGE3 + AGE2
##
## Df Sum of Sq RSS AIC
## + NODENVST 1 39.876 19916 -689841
## + NOEXER 1 37.343 19919 -689807
## + SMOKE 1 31.086 19925 -689723
## + INC1 1 30.943 19925 -689721
## + FRMRSMK 1 27.005 19929 -689669
## + COLLEGE 1 25.754 19930 -689652
## + INC2 1 18.862 19937 -689560
## + LOWED 1 17.141 19939 -689537
## + AGE1 1 13.769 19942 -689492
## + MOSTTEETH 1 11.900 19944 -689467
## + OTHRACE 1 11.228 19945 -689458
## + OBESE 1 8.264 19948 -689419
## + BLACK 1 6.597 19949 -689396
## + ASIAN 1 2.924 19953 -689347
## + UNDWT 1 1.983 19954 -689335
## + SOMECOLL 1 1.962 19954 -689335
## + HISPANIC 1 0.932 19955 -689321
## + OVWT 1 0.818 19955 -689319
## + INC3 1 0.576 19955 -689316
## <none> 19956 -689310
## + INC4 1 0.045 19956 -689309
##
## Step: AIC=-689840.7
## CVDINFR5 ~ NODIABETE + NOTEETH + MALE + FEWTEETH + AGE3 + AGE2 +
## NODENVST
##
## Df Sum of Sq RSS AIC
## + NOEXER 1 28.7129 19887 -690223
## + FRMRSMK 1 28.4372 19888 -690219
## + SMOKE 1 23.3082 19893 -690150
## + INC1 1 22.9302 19893 -690145
## + COLLEGE 1 16.5963 19899 -690060
## + AGE1 1 16.0003 19900 -690053
## + INC2 1 12.9393 19903 -690012
## + LOWED 1 11.5141 19905 -689993
## + OTHRACE 1 9.8245 19906 -689970
## + MOSTTEETH 1 9.7483 19906 -689969
## + BLACK 1 8.5819 19907 -689953
## + OBESE 1 6.8283 19909 -689930
## + ASIAN 1 2.7457 19913 -689875
## + SOMECOLL 1 1.8181 19914 -689863
## + UNDWT 1 1.6514 19914 -689861
## + HISPANIC 1 1.5471 19914 -689859
## + OVWT 1 0.5289 19915 -689846
## + INC3 1 0.2387 19916 -689842
## <none> 19916 -689841
## + INC4 1 0.0507 19916 -689839
##
## Step: AIC=-690222.6
## CVDINFR5 ~ NODIABETE + NOTEETH + MALE + FEWTEETH + AGE3 + AGE2 +
## NODENVST + NOEXER
##
## Df Sum of Sq RSS AIC
## + FRMRSMK 1 28.9327 19858 -690608
## + INC1 1 19.7319 19868 -690485
## + SMOKE 1 19.4796 19868 -690481
## + AGE1 1 16.5024 19871 -690441
## + COLLEGE 1 11.4378 19876 -690374
## + INC2 1 10.7923 19877 -690365
## + OTHRACE 1 9.8141 19877 -690352
## + BLACK 1 9.2235 19878 -690344
## + LOWED 1 8.9543 19878 -690340
## + MOSTTEETH 1 8.2146 19879 -690331
## + OBESE 1 4.2577 19883 -690278
## + ASIAN 1 2.6046 19885 -690255
## + HISPANIC 1 2.1230 19885 -690249
## + SOMECOLL 1 1.7482 19886 -690244
## + UNDWT 1 1.4986 19886 -690241
## + OVWT 1 0.2431 19887 -690224
## <none> 19887 -690223
## + INC3 1 0.1340 19887 -690222
## + INC4 1 0.0443 19887 -690221
##
## Step: AIC=-690608
## CVDINFR5 ~ NODIABETE + NOTEETH + MALE + FEWTEETH + AGE3 + AGE2 +
## NODENVST + NOEXER + FRMRSMK
##
## Df Sum of Sq RSS AIC
## + SMOKE 1 38.012 19820 -691116
## + INC1 1 21.108 19837 -690889
## + AGE1 1 13.849 19845 -690792
## + INC2 1 10.860 19848 -690752
## + COLLEGE 1 10.257 19848 -690743
## + OTHRACE 1 10.096 19848 -690741
## + LOWED 1 9.529 19849 -690734
## + BLACK 1 7.829 19851 -690711
## + MOSTTEETH 1 6.850 19852 -690698
## + OBESE 1 3.479 19855 -690653
## + ASIAN 1 2.196 19856 -690635
## + UNDWT 1 1.704 19857 -690629
## + HISPANIC 1 1.684 19857 -690629
## + SOMECOLL 1 1.250 19857 -690623
## + OVWT 1 0.322 19858 -690610
## <none> 19858 -690608
## + INC3 1 0.100 19858 -690607
## + INC4 1 0.012 19858 -690606
##
## Step: AIC=-691115.9
## CVDINFR5 ~ NODIABETE + NOTEETH + MALE + FEWTEETH + AGE3 + AGE2 +
## NODENVST + NOEXER + FRMRSMK + SMOKE
##
## Df Sum of Sq RSS AIC
## + INC1 1 17.3356 19803 -691347
## + AGE1 1 16.2414 19804 -691332
## + INC2 1 9.0891 19811 -691236
## + OTHRACE 1 8.7990 19812 -691232
## + LOWED 1 8.1048 19812 -691223
## + BLACK 1 7.4285 19813 -691214
## + COLLEGE 1 6.8841 19813 -691206
## + OBESE 1 5.2670 19815 -691185
## + MOSTTEETH 1 4.6946 19816 -691177
## + ASIAN 1 1.8054 19819 -691138
## + HISPANIC 1 1.2279 19819 -691130
## + UNDWT 1 0.9332 19819 -691126
## + SOMECOLL 1 0.8230 19820 -691125
## + OVWT 1 0.3534 19820 -691119
## <none> 19820 -691116
## + INC3 1 0.0346 19820 -691114
## + INC4 1 0.0008 19820 -691114
##
## Step: AIC=-691346.7
## CVDINFR5 ~ NODIABETE + NOTEETH + MALE + FEWTEETH + AGE3 + AGE2 +
## NODENVST + NOEXER + FRMRSMK + SMOKE + INC1
##
## Df Sum of Sq RSS AIC
## + AGE1 1 16.8994 19786 -691572
## + INC2 1 14.7809 19788 -691543
## + BLACK 1 8.8460 19794 -691464
## + OTHRACE 1 7.8240 19795 -691450
## + COLLEGE 1 5.2254 19798 -691415
## + LOWED 1 5.2046 19798 -691415
## + OBESE 1 4.9172 19798 -691411
## + MOSTTEETH 1 4.1730 19799 -691401
## + HISPANIC 1 2.2437 19801 -691375
## + ASIAN 1 1.8125 19801 -691369
## + SOMECOLL 1 1.0270 19802 -691359
## + UNDWT 1 0.7927 19802 -691355
## + INC3 1 0.4411 19803 -691351
## + OVWT 1 0.2884 19803 -691349
## + INC4 1 0.2366 19803 -691348
## <none> 19803 -691347
##
## Step: AIC=-691571.9
## CVDINFR5 ~ NODIABETE + NOTEETH + MALE + FEWTEETH + AGE3 + AGE2 +
## NODENVST + NOEXER + FRMRSMK + SMOKE + INC1 + AGE1
##
## Df Sum of Sq RSS AIC
## + INC2 1 14.4015 19772 -691764
## + BLACK 1 8.4955 19778 -691684
## + OTHRACE 1 8.2307 19778 -691681
## + LOWED 1 5.6971 19780 -691647
## + OBESE 1 5.6198 19781 -691645
## + COLLEGE 1 5.5577 19781 -691645
## + MOSTTEETH 1 3.2422 19783 -691614
## + ASIAN 1 1.6918 19784 -691593
## + HISPANIC 1 1.6346 19784 -691592
## + SOMECOLL 1 0.9289 19785 -691582
## + UNDWT 1 0.7268 19785 -691580
## + OVWT 1 0.3563 19786 -691575
## + INC3 1 0.3198 19786 -691574
## <none> 19786 -691572
## + INC4 1 0.1254 19786 -691572
##
## Step: AIC=-691763.7
## CVDINFR5 ~ NODIABETE + NOTEETH + MALE + FEWTEETH + AGE3 + AGE2 +
## NODENVST + NOEXER + FRMRSMK + SMOKE + INC1 + AGE1 + INC2
##
## Df Sum of Sq RSS AIC
## + BLACK 1 9.4664 19762 -691889
## + OTHRACE 1 7.6466 19764 -691865
## + OBESE 1 5.2308 19767 -691832
## + LOWED 1 4.4231 19767 -691821
## + COLLEGE 1 3.5450 19768 -691809
## + MOSTTEETH 1 2.6249 19769 -691797
## + HISPANIC 1 2.4435 19769 -691795
## + ASIAN 1 1.6820 19770 -691784
## + INC3 1 1.5503 19770 -691783
## + INC4 1 1.0566 19771 -691776
## + SOMECOLL 1 0.8865 19771 -691774
## + UNDWT 1 0.6613 19771 -691771
## + OVWT 1 0.3458 19771 -691766
## <none> 19772 -691764
##
## Step: AIC=-691889.1
## CVDINFR5 ~ NODIABETE + NOTEETH + MALE + FEWTEETH + AGE3 + AGE2 +
## NODENVST + NOEXER + FRMRSMK + SMOKE + INC1 + AGE1 + INC2 +
## BLACK
##
## Df Sum of Sq RSS AIC
## + OTHRACE 1 6.4541 19756 -691974
## + OBESE 1 5.6850 19757 -691964
## + LOWED 1 4.8814 19757 -691953
## + COLLEGE 1 3.7810 19758 -691938
## + HISPANIC 1 3.4135 19759 -691933
## + MOSTTEETH 1 3.1468 19759 -691929
## + ASIAN 1 1.9674 19760 -691914
## + INC3 1 1.7251 19761 -691910
## + INC4 1 1.0967 19761 -691902
## + SOMECOLL 1 0.8383 19761 -691898
## + UNDWT 1 0.6219 19762 -691895
## + OVWT 1 0.3387 19762 -691892
## <none> 19762 -691889
##
## Step: AIC=-691974
## CVDINFR5 ~ NODIABETE + NOTEETH + MALE + FEWTEETH + AGE3 + AGE2 +
## NODENVST + NOEXER + FRMRSMK + SMOKE + INC1 + AGE1 + INC2 +
## BLACK + OTHRACE
##
## Df Sum of Sq RSS AIC
## + OBESE 1 5.5672 19750 -692047
## + LOWED 1 4.7482 19751 -692036
## + COLLEGE 1 3.6460 19752 -692021
## + MOSTTEETH 1 3.0238 19753 -692013
## + HISPANIC 1 2.8174 19753 -692010
## + ASIAN 1 1.7855 19754 -691996
## + INC3 1 1.6394 19754 -691994
## + INC4 1 1.0828 19755 -691987
## + SOMECOLL 1 0.7757 19755 -691982
## + UNDWT 1 0.6135 19755 -691980
## + OVWT 1 0.3266 19755 -691976
## <none> 19756 -691974
##
## Step: AIC=-692047
## CVDINFR5 ~ NODIABETE + NOTEETH + MALE + FEWTEETH + AGE3 + AGE2 +
## NODENVST + NOEXER + FRMRSMK + SMOKE + INC1 + AGE1 + INC2 +
## BLACK + OTHRACE + OBESE
##
## Df Sum of Sq RSS AIC
## + LOWED 1 4.8350 19745 -692110
## + COLLEGE 1 3.3144 19747 -692090
## + HISPANIC 1 2.6823 19748 -692081
## + MOSTTEETH 1 2.6724 19748 -692081
## + INC3 1 1.5229 19749 -692066
## + ASIAN 1 1.4868 19749 -692065
## + INC4 1 0.9485 19749 -692058
## + UNDWT 1 0.8908 19749 -692057
## + SOMECOLL 1 0.6635 19750 -692054
## + OVWT 1 0.4642 19750 -692051
## <none> 19750 -692047
##
## Step: AIC=-692110.2
## CVDINFR5 ~ NODIABETE + NOTEETH + MALE + FEWTEETH + AGE3 + AGE2 +
## NODENVST + NOEXER + FRMRSMK + SMOKE + INC1 + AGE1 + INC2 +
## BLACK + OTHRACE + OBESE + LOWED
##
## Df Sum of Sq RSS AIC
## + HISPANIC 1 4.4496 19741 -692168
## + MOSTTEETH 1 2.5871 19743 -692143
## + COLLEGE 1 2.4173 19743 -692141
## + SOMECOLL 1 1.5039 19744 -692128
## + ASIAN 1 1.4738 19744 -692128
## + INC3 1 1.4580 19744 -692128
## + INC4 1 1.0416 19744 -692122
## + UNDWT 1 0.8757 19745 -692120
## + OVWT 1 0.5002 19745 -692115
## <none> 19745 -692110
##
## Step: AIC=-692168.2
## CVDINFR5 ~ NODIABETE + NOTEETH + MALE + FEWTEETH + AGE3 + AGE2 +
## NODENVST + NOEXER + FRMRSMK + SMOKE + INC1 + AGE1 + INC2 +
## BLACK + OTHRACE + OBESE + LOWED + HISPANIC
##
## Df Sum of Sq RSS AIC
## + MOSTTEETH 1 2.87531 19738 -692205
## + COLLEGE 1 2.59985 19738 -692201
## + ASIAN 1 1.67302 19739 -692189
## + INC3 1 1.64521 19739 -692188
## + SOMECOLL 1 1.53671 19739 -692187
## + INC4 1 1.07560 19740 -692181
## + UNDWT 1 0.83990 19740 -692177
## + OVWT 1 0.52782 19740 -692173
## <none> 19741 -692168
##
## Step: AIC=-692204.9
## CVDINFR5 ~ NODIABETE + NOTEETH + MALE + FEWTEETH + AGE3 + AGE2 +
## NODENVST + NOEXER + FRMRSMK + SMOKE + INC1 + AGE1 + INC2 +
## BLACK + OTHRACE + OBESE + LOWED + HISPANIC + MOSTTEETH
##
## Df Sum of Sq RSS AIC
## + COLLEGE 1 2.12124 19736 -692232
## + ASIAN 1 1.71834 19736 -692226
## + INC3 1 1.45601 19737 -692223
## + SOMECOLL 1 1.41067 19737 -692222
## + INC4 1 0.93873 19737 -692216
## + UNDWT 1 0.85952 19737 -692215
## + OVWT 1 0.46493 19738 -692209
## <none> 19738 -692205
##
## Step: AIC=-692231.5
## CVDINFR5 ~ NODIABETE + NOTEETH + MALE + FEWTEETH + AGE3 + AGE2 +
## NODENVST + NOEXER + FRMRSMK + SMOKE + INC1 + AGE1 + INC2 +
## BLACK + OTHRACE + OBESE + LOWED + HISPANIC + MOSTTEETH +
## COLLEGE
##
## Df Sum of Sq RSS AIC
## + ASIAN 1 1.57968 19734 -692251
## + INC3 1 1.14339 19735 -692245
## + UNDWT 1 0.87654 19735 -692241
## + INC4 1 0.74951 19735 -692240
## + OVWT 1 0.41931 19736 -692235
## + SOMECOLL 1 0.22718 19736 -692233
## <none> 19736 -692232
##
## Step: AIC=-692250.8
## CVDINFR5 ~ NODIABETE + NOTEETH + MALE + FEWTEETH + AGE3 + AGE2 +
## NODENVST + NOEXER + FRMRSMK + SMOKE + INC1 + AGE1 + INC2 +
## BLACK + OTHRACE + OBESE + LOWED + HISPANIC + MOSTTEETH +
## COLLEGE + ASIAN
##
## Df Sum of Sq RSS AIC
## + INC3 1 1.16526 19733 -692265
## + UNDWT 1 0.91539 19733 -692261
## + INC4 1 0.74986 19734 -692259
## + OVWT 1 0.37003 19734 -692254
## + SOMECOLL 1 0.23341 19734 -692252
## <none> 19734 -692251
##
## Step: AIC=-692264.5
## CVDINFR5 ~ NODIABETE + NOTEETH + MALE + FEWTEETH + AGE3 + AGE2 +
## NODENVST + NOEXER + FRMRSMK + SMOKE + INC1 + AGE1 + INC2 +
## BLACK + OTHRACE + OBESE + LOWED + HISPANIC + MOSTTEETH +
## COLLEGE + ASIAN + INC3
##
## Df Sum of Sq RSS AIC
## + INC4 1 1.16058 19732 -692278
## + UNDWT 1 0.90064 19732 -692275
## + OVWT 1 0.35668 19733 -692267
## + SOMECOLL 1 0.25724 19733 -692266
## <none> 19733 -692265
##
## Step: AIC=-692278.2
## CVDINFR5 ~ NODIABETE + NOTEETH + MALE + FEWTEETH + AGE3 + AGE2 +
## NODENVST + NOEXER + FRMRSMK + SMOKE + INC1 + AGE1 + INC2 +
## BLACK + OTHRACE + OBESE + LOWED + HISPANIC + MOSTTEETH +
## COLLEGE + ASIAN + INC3 + INC4
##
## Df Sum of Sq RSS AIC
## + UNDWT 1 0.90185 19731 -692288
## + OVWT 1 0.32603 19732 -692281
## + SOMECOLL 1 0.25934 19732 -692280
## <none> 19732 -692278
##
## Step: AIC=-692288.3
## CVDINFR5 ~ NODIABETE + NOTEETH + MALE + FEWTEETH + AGE3 + AGE2 +
## NODENVST + NOEXER + FRMRSMK + SMOKE + INC1 + AGE1 + INC2 +
## BLACK + OTHRACE + OBESE + LOWED + HISPANIC + MOSTTEETH +
## COLLEGE + ASIAN + INC3 + INC4 + UNDWT
##
## Df Sum of Sq RSS AIC
## + OVWT 1 0.49761 19731 -692293
## + SOMECOLL 1 0.25699 19731 -692290
## <none> 19731 -692288
##
## Step: AIC=-692293.1
## CVDINFR5 ~ NODIABETE + NOTEETH + MALE + FEWTEETH + AGE3 + AGE2 +
## NODENVST + NOEXER + FRMRSMK + SMOKE + INC1 + AGE1 + INC2 +
## BLACK + OTHRACE + OBESE + LOWED + HISPANIC + MOSTTEETH +
## COLLEGE + ASIAN + INC3 + INC4 + UNDWT + OVWT
##
## Df Sum of Sq RSS AIC
## + SOMECOLL 1 0.25392 19730 -692294
## <none> 19731 -692293
##
## Step: AIC=-692294.5
## CVDINFR5 ~ NODIABETE + NOTEETH + MALE + FEWTEETH + AGE3 + AGE2 +
## NODENVST + NOEXER + FRMRSMK + SMOKE + INC1 + AGE1 + INC2 +
## BLACK + OTHRACE + OBESE + LOWED + HISPANIC + MOSTTEETH +
## COLLEGE + ASIAN + INC3 + INC4 + UNDWT + OVWT + SOMECOLL
##
##
## > summary(fwd.model)
##
## Call:
## lm(formula = CVDINFR5 ~ NODIABETE + NOTEETH + MALE + FEWTEETH +
## AGE3 + AGE2 + NODENVST + NOEXER + FRMRSMK + SMOKE + INC1 +
## AGE1 + INC2 + BLACK + OTHRACE + OBESE + LOWED + HISPANIC +
## MOSTTEETH + COLLEGE + ASIAN + INC3 + INC4 + UNDWT + OVWT +
## SOMECOLL, data = BRFSS)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.37469 -0.11897 -0.06565 -0.01636 1.05550
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.039723 0.002339 16.985 < 2e-16 ***
## NODIABETE -0.069073 0.001407 -49.103 < 2e-16 ***
## NOTEETH 0.062915 0.002116 29.726 < 2e-16 ***
## MALE 0.057156 0.001099 51.991 < 2e-16 ***
## FEWTEETH 0.035902 0.001673 21.462 < 2e-16 ***
## AGE3 0.083418 0.001918 43.483 < 2e-16 ***
## AGE2 0.049538 0.001512 32.759 < 2e-16 ***
## NODENVST 0.016985 0.001288 13.192 < 2e-16 ***
## NOEXER 0.017406 0.001244 13.996 < 2e-16 ***
## FRMRSMK 0.026789 0.001199 22.348 < 2e-16 ***
## SMOKE 0.034479 0.001793 19.227 < 2e-16 ***
## INC1 0.037064 0.002141 17.310 < 2e-16 ***
## AGE1 0.019320 0.001357 14.234 < 2e-16 ***
## INC2 0.023720 0.001706 13.900 < 2e-16 ***
## BLACK -0.025684 0.002053 -12.513 < 2e-16 ***
## OTHRACE 0.021897 0.002682 8.163 3.28e-16 ***
## OBESE 0.011321 0.001393 8.126 4.45e-16 ***
## LOWED 0.020631 0.002272 9.082 < 2e-16 ***
## HISPANIC -0.021033 0.002501 -8.408 < 2e-16 ***
## MOSTTEETH 0.006902 0.001269 5.440 5.32e-08 ***
## COLLEGE -0.003831 0.001433 -2.673 0.007522 **
## ASIAN -0.021501 0.004633 -4.641 3.47e-06 ***
## INC3 0.008899 0.001941 4.585 4.55e-06 ***
## INC4 0.006672 0.001712 3.897 9.74e-05 ***
## UNDWT 0.017320 0.004559 3.799 0.000145 ***
## OVWT 0.003361 0.001301 2.583 0.009809 **
## SOMECOLL 0.002666 0.001441 1.850 0.064246 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.2723 on 266082 degrees of freedom
## Multiple R-squared: 0.06244, Adjusted R-squared: 0.06235
## F-statistic: 681.6 on 26 and 266082 DF, p-value: < 2.2e-16
##
##
## > full.model <- lm(CVDINFR5 ~ NOTEETH + FEWTEETH + MOSTTEETH +
## + MALE + BLACK + HISPANIC + ASIAN + OTHRACE + AGE1 + AGE2 +
## + AGE3 + LOWED + .... [TRUNCATED]
##
## > summary(full.model)
##
## Call:
## lm(formula = CVDINFR5 ~ NOTEETH + FEWTEETH + MOSTTEETH + MALE +
## BLACK + HISPANIC + ASIAN + OTHRACE + AGE1 + AGE2 + AGE3 +
## LOWED + SOMECOLL + COLLEGE + INC1 + INC2 + INC3 + INC4 +
## NODENVST + NODIABETE + FRMRSMK + SMOKE + NOEXER + UNDWT +
## OVWT + OBESE, data = BRFSS)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.37469 -0.11897 -0.06565 -0.01636 1.05550
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.039723 0.002339 16.985 < 2e-16 ***
## NOTEETH 0.062915 0.002116 29.726 < 2e-16 ***
## FEWTEETH 0.035902 0.001673 21.462 < 2e-16 ***
## MOSTTEETH 0.006902 0.001269 5.440 5.32e-08 ***
## MALE 0.057156 0.001099 51.991 < 2e-16 ***
## BLACK -0.025684 0.002053 -12.513 < 2e-16 ***
## HISPANIC -0.021033 0.002501 -8.408 < 2e-16 ***
## ASIAN -0.021501 0.004633 -4.641 3.47e-06 ***
## OTHRACE 0.021897 0.002682 8.163 3.28e-16 ***
## AGE1 0.019320 0.001357 14.234 < 2e-16 ***
## AGE2 0.049538 0.001512 32.759 < 2e-16 ***
## AGE3 0.083418 0.001918 43.483 < 2e-16 ***
## LOWED 0.020631 0.002272 9.082 < 2e-16 ***
## SOMECOLL 0.002666 0.001441 1.850 0.064246 .
## COLLEGE -0.003831 0.001433 -2.673 0.007522 **
## INC1 0.037064 0.002141 17.310 < 2e-16 ***
## INC2 0.023720 0.001706 13.900 < 2e-16 ***
## INC3 0.008899 0.001941 4.585 4.55e-06 ***
## INC4 0.006672 0.001712 3.897 9.74e-05 ***
## NODENVST 0.016985 0.001288 13.192 < 2e-16 ***
## NODIABETE -0.069073 0.001407 -49.103 < 2e-16 ***
## FRMRSMK 0.026789 0.001199 22.348 < 2e-16 ***
## SMOKE 0.034479 0.001793 19.227 < 2e-16 ***
## NOEXER 0.017406 0.001244 13.996 < 2e-16 ***
## UNDWT 0.017320 0.004559 3.799 0.000145 ***
## OVWT 0.003361 0.001301 2.583 0.009809 **
## OBESE 0.011321 0.001393 8.126 4.45e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.2723 on 266082 degrees of freedom
## Multiple R-squared: 0.06244, Adjusted R-squared: 0.06235
## F-statistic: 681.6 on 26 and 266082 DF, p-value: < 2.2e-16
##
##
## > tidy(model1)
## # A tibble: 4 x 5
## term estimate std.error statistic p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 0.0508 0.000859 59.2 0.
## 2 NOTEETH 0.130 0.00190 68.2 0.
## 3 FEWTEETH 0.0823 0.00158 52.3 0.
## 4 MOSTTEETH 0.0264 0.00126 20.9 4.11e-97
##
## > tidy(model2)
## # A tibble: 8 x 5
## term estimate std.error statistic p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 0.000404 0.00124 0.326 7.44e- 1
## 2 NOTEETH 0.115 0.00191 60.1 0.
## 3 FEWTEETH 0.0696 0.00158 44.1 0.
## 4 MOSTTEETH 0.0190 0.00126 15.1 2.15e- 51
## 5 MALE 0.0617 0.00108 57.2 0.
## 6 AGE1 0.0206 0.00136 15.2 4.65e- 52
## 7 AGE2 0.0514 0.00148 34.7 5.76e-263
## 8 AGE3 0.0810 0.00186 43.5 0.
##
## > tidy(full.model)
## # A tibble: 27 x 5
## term estimate std.error statistic p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 0.0397 0.00234 17.0 1.15e- 64
## 2 NOTEETH 0.0629 0.00212 29.7 7.31e-194
## 3 FEWTEETH 0.0359 0.00167 21.5 4.28e-102
## 4 MOSTTEETH 0.00690 0.00127 5.44 5.32e- 8
## 5 MALE 0.0572 0.00110 52.0 0.
## 6 BLACK -0.0257 0.00205 -12.5 6.45e- 36
## 7 HISPANIC -0.0210 0.00250 -8.41 4.18e- 17
## 8 ASIAN -0.0215 0.00463 -4.64 3.47e- 6
## 9 OTHRACE 0.0219 0.00268 8.16 3.28e- 16
## 10 AGE1 0.0193 0.00136 14.2 5.83e- 46
## # ... with 17 more rows
##
## > tidy(fwd.model)
## # A tibble: 27 x 5
## term estimate std.error statistic p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 0.0397 0.00234 17.0 1.15e- 64
## 2 NODIABETE -0.0691 0.00141 -49.1 0.
## 3 NOTEETH 0.0629 0.00212 29.7 7.31e-194
## 4 MALE 0.0572 0.00110 52.0 0.
## 5 FEWTEETH 0.0359 0.00167 21.5 4.28e-102
## 6 AGE3 0.0834 0.00192 43.5 0.
## 7 AGE2 0.0495 0.00151 32.8 6.73e-235
## 8 NODENVST 0.0170 0.00129 13.2 1.01e- 39
## 9 NOEXER 0.0174 0.00124 14.0 1.71e- 44
## 10 FRMRSMK 0.0268 0.00120 22.3 1.61e-110
## # ... with 17 more rows
##
## > write.csv(tidy(model1), file = "../output/linear/TidyRegressionModel1.csv")
##
## > write.csv(tidy(model2), file = "../output/linear/TidyRegressionModel2.csv")
##
## > write.csv(tidy(full.model), file = "../output/linear/TidyRegressionModelFull.csv")
##
## > write.csv(tidy(fwd.model), file = "../output/linear/TidyRegressionModelFWD.csv")
##
## > percent(summary(model1)$adj.r.squared)
## [1] "2.26%"
##
## > percent(summary(model2)$adj.r.squared)
## [1] "4.19%"
##
## > percent(summary(full.model)$adj.r.squared)
## [1] "6.23%"
##
## > summary(model1)$adj.r.squared
## [1] 0.0225682
##
## > summary(model2)$adj.r.squared
## [1] 0.04190584
##
## > summary(full.model)$adj.r.squared
## [1] 0.06234753
##
## > dp1 <- autoplot(model1, label.size = 3) + theme_minimal()
##
## > ggsave(file = "../output/figures/lmodel1diagnostics.png",
## + dp1)
## Saving 7 x 5 in image
##
## > dp2 <- autoplot(model2, label.size = 3) + theme_minimal()
##
## > ggsave(file = "../output/figures/lmodel2diagnostics.png",
## + dp2)
## Saving 7 x 5 in image
##
## > dp3 <- autoplot(full.model, label.size = 3) + theme_minimal()
##
## > ggsave(file = "../output/figures/lmfulldiagnostics.png",
## + dp3)
## Saving 7 x 5 in image
##
## > gvmodel <- gvlma(model1)
##
## > summary(gvmodel)
##
## Call:
## lm(formula = CVDINFR5 ~ NOTEETH + FEWTEETH + MOSTTEETH, data = BRFSS)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.18069 -0.07726 -0.07726 -0.05084 0.94916
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.0508387 0.0008588 59.20 <2e-16 ***
## NOTEETH 0.1298464 0.0019032 68.23 <2e-16 ***
## FEWTEETH 0.0823156 0.0015754 52.25 <2e-16 ***
## MOSTTEETH 0.0264187 0.0012628 20.92 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.278 on 266105 degrees of freedom
## Multiple R-squared: 0.02258, Adjusted R-squared: 0.02257
## F-statistic: 2049 on 3 and 266105 DF, p-value: < 2.2e-16
##
##
## ASSESSMENT OF THE LINEAR MODEL ASSUMPTIONS
## USING THE GLOBAL TEST ON 4 DEGREES-OF-FREEDOM:
## Level of Significance = 0.05
##
## Call:
## gvlma(x = model1)
##
## Value p-value Decision
## Global Stat 8.115e+05 0.000e+00 Assumptions NOT satisfied!
## Skewness 3.588e+05 0.000e+00 Assumptions NOT satisfied!
## Kurtosis 4.527e+05 0.000e+00 Assumptions NOT satisfied!
## Link Function 8.712e-14 1.000e+00 Assumptions acceptable.
## Heteroscedasticity 2.240e+01 2.212e-06 Assumptions NOT satisfied!
##
## > gvmodel <- gvlma(model2)
##
## > summary(gvmodel)
##
## Call:
## lm(formula = CVDINFR5 ~ NOTEETH + FEWTEETH + MOSTTEETH + MALE +
## AGE1 + AGE2 + AGE3, data = BRFSS)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.25791 -0.11350 -0.07078 -0.02102 0.99960
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.0004043 0.0012393 0.326 0.744
## NOTEETH 0.1148229 0.0019111 60.083 <2e-16 ***
## FEWTEETH 0.0695639 0.0015779 44.086 <2e-16 ***
## MOSTTEETH 0.0189619 0.0012570 15.084 <2e-16 ***
## MALE 0.0616769 0.0010787 57.177 <2e-16 ***
## AGE1 0.0206114 0.0013573 15.185 <2e-16 ***
## AGE2 0.0514180 0.0014826 34.682 <2e-16 ***
## AGE3 0.0810020 0.0018611 43.523 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.2753 on 266101 degrees of freedom
## Multiple R-squared: 0.04193, Adjusted R-squared: 0.04191
## F-statistic: 1664 on 7 and 266101 DF, p-value: < 2.2e-16
##
##
## ASSESSMENT OF THE LINEAR MODEL ASSUMPTIONS
## USING THE GLOBAL TEST ON 4 DEGREES-OF-FREEDOM:
## Level of Significance = 0.05
##
## Call:
## gvlma(x = model2)
##
## Value p-value Decision
## Global Stat 751733.62 0.000e+00 Assumptions NOT satisfied!
## Skewness 336638.98 0.000e+00 Assumptions NOT satisfied!
## Kurtosis 414811.49 0.000e+00 Assumptions NOT satisfied!
## Link Function 262.41 0.000e+00 Assumptions NOT satisfied!
## Heteroscedasticity 20.74 5.254e-06 Assumptions NOT satisfied!
##
## > gvmodel <- gvlma(full.model)
##
## > summary(gvmodel)
##
## Call:
## lm(formula = CVDINFR5 ~ NOTEETH + FEWTEETH + MOSTTEETH + MALE +
## BLACK + HISPANIC + ASIAN + OTHRACE + AGE1 + AGE2 + AGE3 +
## LOWED + SOMECOLL + COLLEGE + INC1 + INC2 + INC3 + INC4 +
## NODENVST + NODIABETE + FRMRSMK + SMOKE + NOEXER + UNDWT +
## OVWT + OBESE, data = BRFSS)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.37469 -0.11897 -0.06565 -0.01636 1.05550
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.039723 0.002339 16.985 < 2e-16 ***
## NOTEETH 0.062915 0.002116 29.726 < 2e-16 ***
## FEWTEETH 0.035902 0.001673 21.462 < 2e-16 ***
## MOSTTEETH 0.006902 0.001269 5.440 5.32e-08 ***
## MALE 0.057156 0.001099 51.991 < 2e-16 ***
## BLACK -0.025684 0.002053 -12.513 < 2e-16 ***
## HISPANIC -0.021033 0.002501 -8.408 < 2e-16 ***
## ASIAN -0.021501 0.004633 -4.641 3.47e-06 ***
## OTHRACE 0.021897 0.002682 8.163 3.28e-16 ***
## AGE1 0.019320 0.001357 14.234 < 2e-16 ***
## AGE2 0.049538 0.001512 32.759 < 2e-16 ***
## AGE3 0.083418 0.001918 43.483 < 2e-16 ***
## LOWED 0.020631 0.002272 9.082 < 2e-16 ***
## SOMECOLL 0.002666 0.001441 1.850 0.064246 .
## COLLEGE -0.003831 0.001433 -2.673 0.007522 **
## INC1 0.037064 0.002141 17.310 < 2e-16 ***
## INC2 0.023720 0.001706 13.900 < 2e-16 ***
## INC3 0.008899 0.001941 4.585 4.55e-06 ***
## INC4 0.006672 0.001712 3.897 9.74e-05 ***
## NODENVST 0.016985 0.001288 13.192 < 2e-16 ***
## NODIABETE -0.069073 0.001407 -49.103 < 2e-16 ***
## FRMRSMK 0.026789 0.001199 22.348 < 2e-16 ***
## SMOKE 0.034479 0.001793 19.227 < 2e-16 ***
## NOEXER 0.017406 0.001244 13.996 < 2e-16 ***
## UNDWT 0.017320 0.004559 3.799 0.000145 ***
## OVWT 0.003361 0.001301 2.583 0.009809 **
## OBESE 0.011321 0.001393 8.126 4.45e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.2723 on 266082 degrees of freedom
## Multiple R-squared: 0.06244, Adjusted R-squared: 0.06235
## F-statistic: 681.6 on 26 and 266082 DF, p-value: < 2.2e-16
##
##
## ASSESSMENT OF THE LINEAR MODEL ASSUMPTIONS
## USING THE GLOBAL TEST ON 4 DEGREES-OF-FREEDOM:
## Level of Significance = 0.05
##
## Call:
## gvlma(x = full.model)
##
## Value p-value Decision
## Global Stat 694863.46 0.000e+00 Assumptions NOT satisfied!
## Skewness 314383.05 0.000e+00 Assumptions NOT satisfied!
## Kurtosis 379489.65 0.000e+00 Assumptions NOT satisfied!
## Link Function 969.92 0.000e+00 Assumptions NOT satisfied!
## Heteroscedasticity 20.84 4.984e-06 Assumptions NOT satisfied!
Model 1 Diagnostics
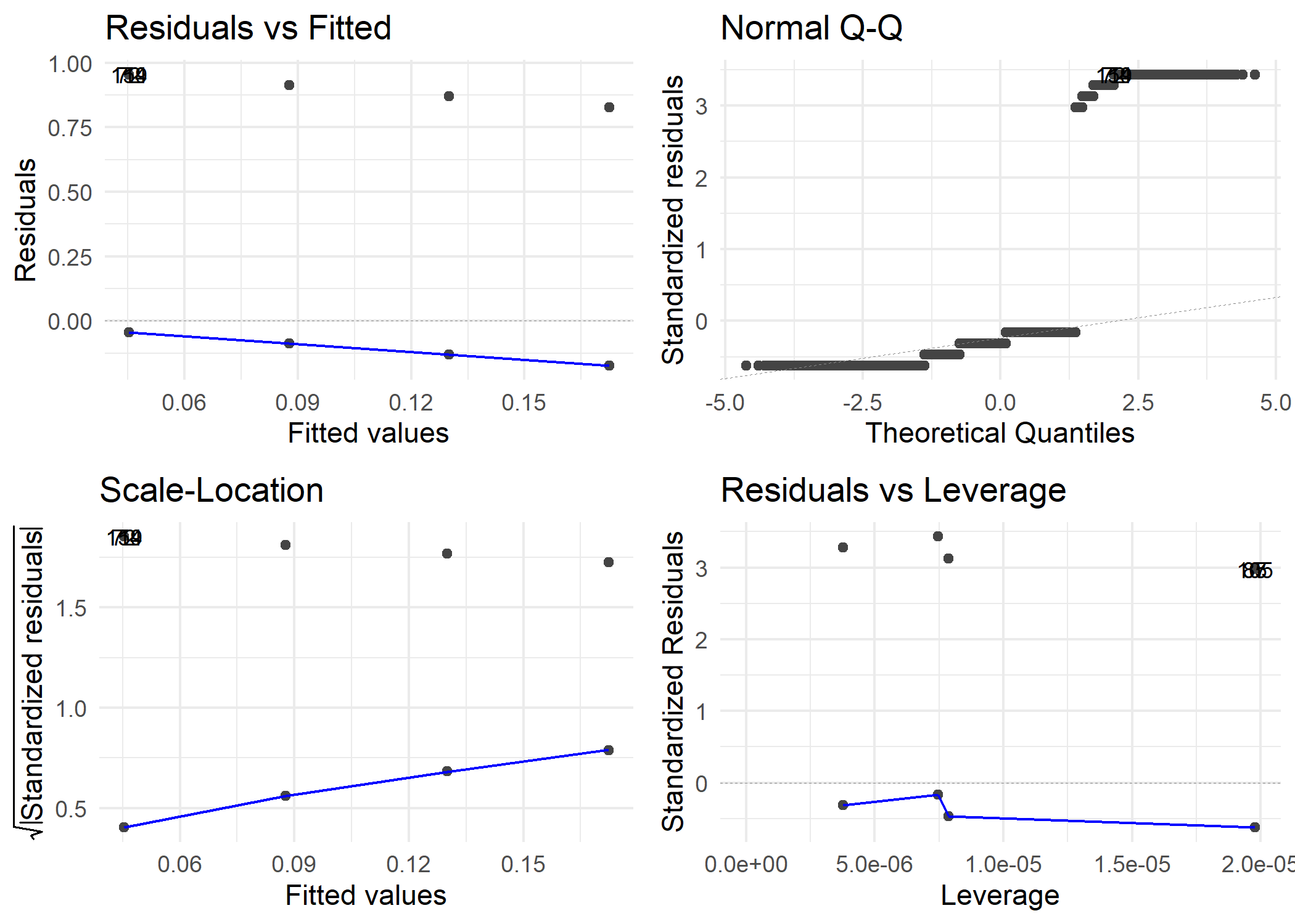
Model 2 Diagnostics
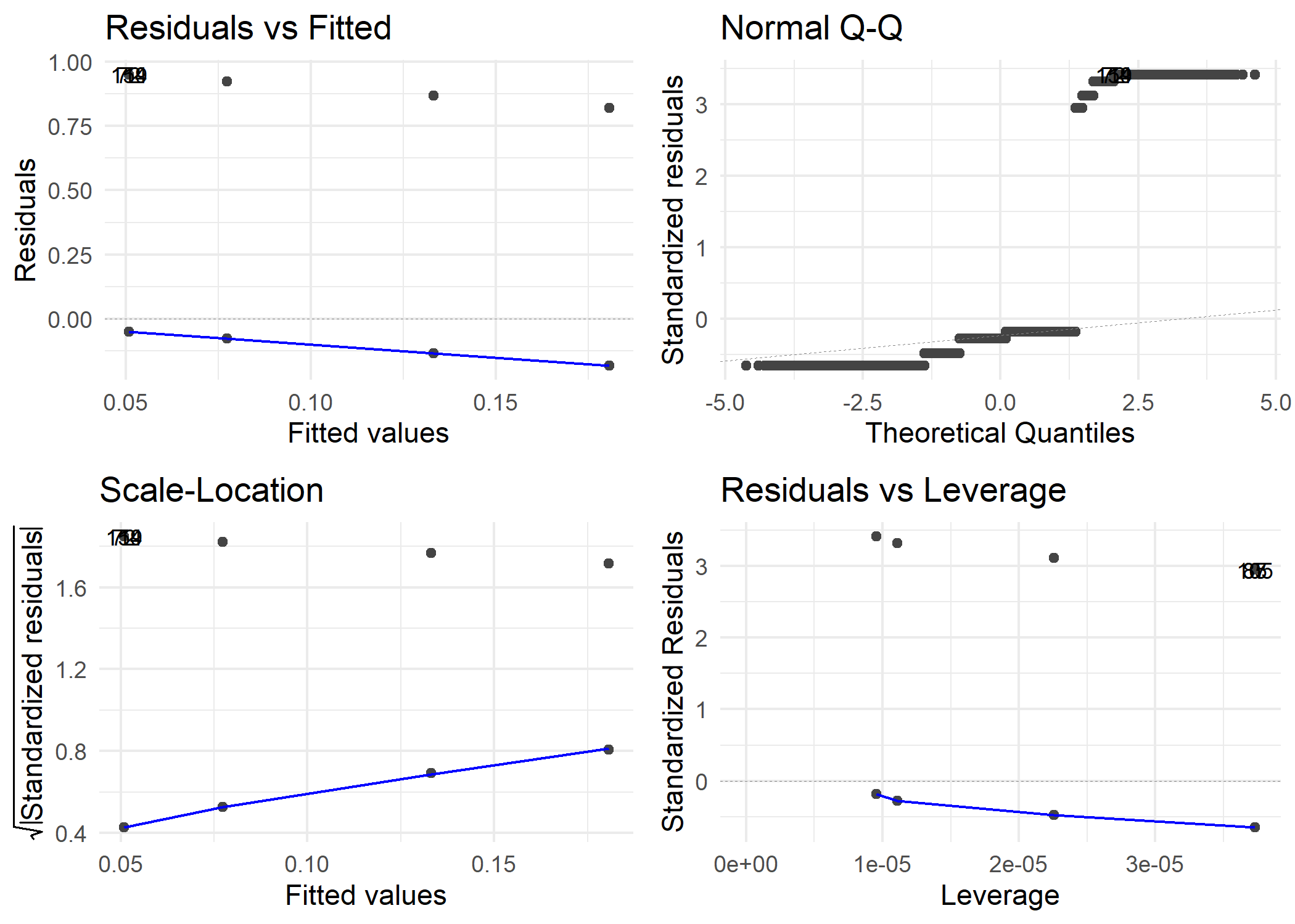
Full Model Diagnostics
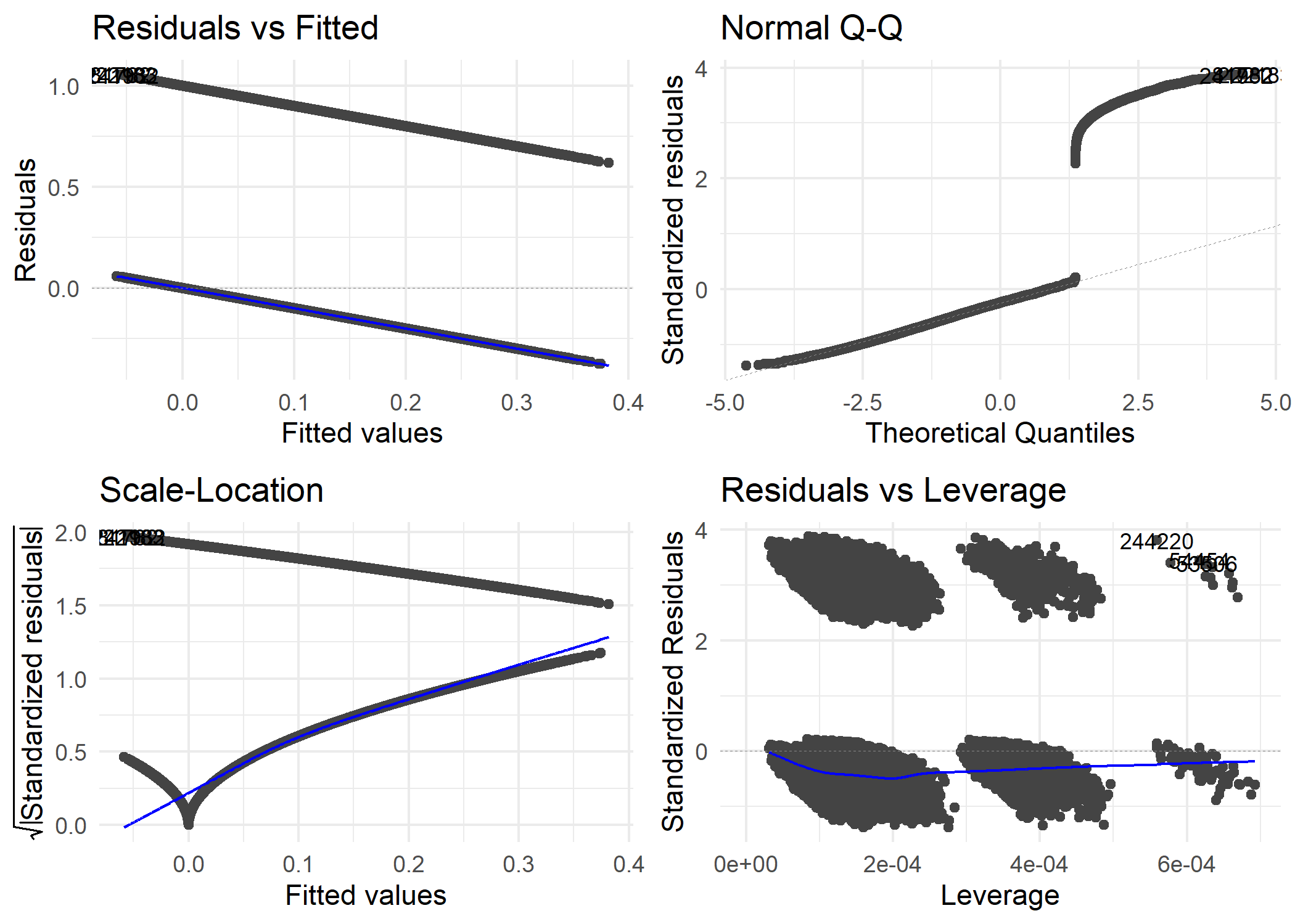
This concluded the linear regression modeling.
Return to the Main Overview