Logistic Regression
First created on Nov 8, 2019. Updated on Nov 11, 2019
Step 1: Load Libraries
source("../code/100_Dependencies.R", echo = TRUE)
##
## > library(foreign)
##
## > library(Hmisc)
## Loading required package: lattice
## Loading required package: survival
## Loading required package: Formula
## Loading required package: ggplot2
##
## Attaching package: 'Hmisc'
## The following objects are masked from 'package:base':
##
## format.pval, units
##
## > library(e1071)
##
## Attaching package: 'e1071'
## The following object is masked from 'package:Hmisc':
##
## impute
##
## > library(gtools)
##
## Attaching package: 'gtools'
## The following object is masked from 'package:e1071':
##
## permutations
##
## > library(MASS)
##
## > library(ggplot2)
##
## > library(gridExtra)
##
## > library(broom)
##
## > library(scales)
##
## > library(ggfortify)
##
## > library(gvlma)
Step 2: Run Logistic Regression Models
source("../code/265_Logistic regression models.R", echo = TRUE)
##
## > BRFSS <- read.csv(file = "../output/BRFSS.csv", header = TRUE,
## + sep = ",")
##
## > logModel1 <- glm(CVDINFR5 ~ NOTEETH + FEWTEETH + MOSTTEETH,
## + data = BRFSS, family = "binomial")
##
## > summary(logModel1)
##
## Call:
## glm(formula = CVDINFR5 ~ NOTEETH + FEWTEETH + MOSTTEETH, family = "binomial",
## data = BRFSS)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -0.6313 -0.4010 -0.4010 -0.3230 2.4409
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -2.92692 0.01406 -208.15 <2e-16 ***
## NOTEETH 1.41521 0.02121 66.73 <2e-16 ***
## FEWTEETH 1.05357 0.01983 53.13 <2e-16 ***
## MOSTTEETH 0.44671 0.01879 23.77 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 156763 on 266108 degrees of freedom
## Residual deviance: 151289 on 266105 degrees of freedom
## AIC: 151297
##
## Number of Fisher Scoring iterations: 5
##
##
## > logModel2 <- glm(CVDINFR5 ~ NOTEETH + FEWTEETH + MOSTTEETH +
## + MALE + AGE1 + AGE2 + AGE3, data = BRFSS, family = "binomial")
##
## > summary(logModel2)
##
## Call:
## glm(formula = CVDINFR5 ~ NOTEETH + FEWTEETH + MOSTTEETH + MALE +
## AGE1 + AGE2 + AGE3, family = "binomial", data = BRFSS)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -0.9099 -0.4594 -0.3699 -0.2593 2.7490
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -3.75548 0.02187 -171.72 <2e-16 ***
## NOTEETH 1.25509 0.02180 57.59 <2e-16 ***
## FEWTEETH 0.90374 0.02024 44.64 <2e-16 ***
## MOSTTEETH 0.35155 0.01902 18.49 <2e-16 ***
## MALE 0.80400 0.01439 55.88 <2e-16 ***
## AGE1 0.37972 0.02115 17.95 <2e-16 ***
## AGE2 0.75589 0.02123 35.60 <2e-16 ***
## AGE3 1.02837 0.02377 43.27 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 156763 on 266108 degrees of freedom
## Residual deviance: 146057 on 266101 degrees of freedom
## AIC: 146073
##
## Number of Fisher Scoring iterations: 5
##
##
## > logFullModel <- glm(CVDINFR5 ~ NOTEETH + FEWTEETH +
## + MOSTTEETH + MALE + BLACK + HISPANIC + ASIAN + OTHRACE + AGE1 +
## + AGE2 + AGE3 + LOWED .... [TRUNCATED]
##
## > summary(logFullModel)
##
## Call:
## glm(formula = CVDINFR5 ~ NOTEETH + FEWTEETH + MOSTTEETH + MALE +
## BLACK + HISPANIC + ASIAN + OTHRACE + AGE1 + AGE2 + AGE3 +
## LOWED + SOMECOLL + COLLEGE + INC1 + INC2 + INC3 + INC4 +
## NODENVST + NODIABETE + FRMRSMK + SMOKE + NOEXER + UNDWT +
## OVWT + OBESE, family = "binomial", data = BRFSS)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -1.4537 -0.4532 -0.3282 -0.2347 3.0642
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -3.57592 0.03386 -105.615 < 2e-16 ***
## NOTEETH 0.62128 0.02476 25.090 < 2e-16 ***
## FEWTEETH 0.49110 0.02160 22.740 < 2e-16 ***
## MOSTTEETH 0.20154 0.01942 10.380 < 2e-16 ***
## MALE 0.77617 0.01504 51.618 < 2e-16 ***
## BLACK -0.30616 0.02853 -10.731 < 2e-16 ***
## HISPANIC -0.23536 0.03543 -6.643 3.08e-11 ***
## ASIAN -0.34816 0.07742 -4.497 6.88e-06 ***
## OTHRACE 0.25326 0.03114 8.134 4.17e-16 ***
## AGE1 0.39748 0.02163 18.373 < 2e-16 ***
## AGE2 0.80548 0.02227 36.162 < 2e-16 ***
## AGE3 1.16911 0.02541 46.007 < 2e-16 ***
## LOWED 0.16342 0.02517 6.492 8.46e-11 ***
## SOMECOLL 0.02251 0.01865 1.207 0.227
## COLLEGE -0.13705 0.02008 -6.825 8.79e-12 ***
## INC1 0.45549 0.02476 18.396 < 2e-16 ***
## INC2 0.30338 0.02085 14.547 < 2e-16 ***
## INC3 0.13739 0.02503 5.490 4.03e-08 ***
## INC4 0.10841 0.02327 4.658 3.20e-06 ***
## NODENVST 0.21640 0.01634 13.243 < 2e-16 ***
## NODIABETE -0.70871 0.01594 -44.462 < 2e-16 ***
## FRMRSMK 0.35038 0.01616 21.676 < 2e-16 ***
## SMOKE 0.51351 0.02246 22.866 < 2e-16 ***
## NOEXER 0.22572 0.01550 14.563 < 2e-16 ***
## UNDWT 0.25637 0.05710 4.490 7.14e-06 ***
## OVWT 0.08429 0.01845 4.569 4.90e-06 ***
## OBESE 0.20773 0.01913 10.856 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 156763 on 266108 degrees of freedom
## Residual deviance: 140689 on 266082 degrees of freedom
## AIC: 140743
##
## Number of Fisher Scoring iterations: 6
##
##
## > stepLogModel <- stepAIC(logFullModel)
## Start: AIC=140743.2
## CVDINFR5 ~ NOTEETH + FEWTEETH + MOSTTEETH + MALE + BLACK + HISPANIC +
## ASIAN + OTHRACE + AGE1 + AGE2 + AGE3 + LOWED + SOMECOLL +
## COLLEGE + INC1 + INC2 + INC3 + INC4 + NODENVST + NODIABETE +
## FRMRSMK + SMOKE + NOEXER + UNDWT + OVWT + OBESE
##
## Df Deviance AIC
## - SOMECOLL 1 140691 140743
## <none> 140689 140743
## - UNDWT 1 140708 140760
## - OVWT 1 140710 140762
## - INC4 1 140711 140763
## - ASIAN 1 140711 140763
## - INC3 1 140719 140771
## - LOWED 1 140731 140783
## - HISPANIC 1 140735 140787
## - COLLEGE 1 140736 140788
## - OTHRACE 1 140752 140804
## - MOSTTEETH 1 140798 140850
## - OBESE 1 140808 140860
## - BLACK 1 140811 140863
## - NODENVST 1 140863 140915
## - INC2 1 140896 140948
## - NOEXER 1 140899 140951
## - INC1 1 141014 141066
## - AGE1 1 141036 141088
## - FRMRSMK 1 141158 141210
## - SMOKE 1 141192 141244
## - FEWTEETH 1 141205 141257
## - NOTEETH 1 141313 141365
## - AGE2 1 142060 142112
## - NODIABETE 1 142576 142628
## - AGE3 1 142819 142871
## - MALE 1 143424 143476
##
## Step: AIC=140742.7
## CVDINFR5 ~ NOTEETH + FEWTEETH + MOSTTEETH + MALE + BLACK + HISPANIC +
## ASIAN + OTHRACE + AGE1 + AGE2 + AGE3 + LOWED + COLLEGE +
## INC1 + INC2 + INC3 + INC4 + NODENVST + NODIABETE + FRMRSMK +
## SMOKE + NOEXER + UNDWT + OVWT + OBESE
##
## Df Deviance AIC
## <none> 140691 140743
## - UNDWT 1 140710 140760
## - OVWT 1 140712 140762
## - INC4 1 140712 140762
## - ASIAN 1 140713 140763
## - INC3 1 140720 140770
## - LOWED 1 140731 140781
## - HISPANIC 1 140737 140787
## - OTHRACE 1 140754 140804
## - COLLEGE 1 140761 140811
## - MOSTTEETH 1 140799 140849
## - OBESE 1 140809 140859
## - BLACK 1 140813 140863
## - NODENVST 1 140863 140913
## - INC2 1 140896 140946
## - NOEXER 1 140899 140949
## - INC1 1 141014 141064
## - AGE1 1 141039 141089
## - FRMRSMK 1 141160 141210
## - SMOKE 1 141194 141244
## - FEWTEETH 1 141205 141255
## - NOTEETH 1 141313 141363
## - AGE2 1 142063 142113
## - NODIABETE 1 142578 142628
## - AGE3 1 142819 142869
## - MALE 1 143424 143474
##
## > summary(stepLogModel)
##
## Call:
## glm(formula = CVDINFR5 ~ NOTEETH + FEWTEETH + MOSTTEETH + MALE +
## BLACK + HISPANIC + ASIAN + OTHRACE + AGE1 + AGE2 + AGE3 +
## LOWED + COLLEGE + INC1 + INC2 + INC3 + INC4 + NODENVST +
## NODIABETE + FRMRSMK + SMOKE + NOEXER + UNDWT + OVWT + OBESE,
## family = "binomial", data = BRFSS)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -1.4532 -0.4530 -0.3280 -0.2351 3.0639
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -3.56371 0.03230 -110.323 < 2e-16 ***
## NOTEETH 0.61946 0.02471 25.064 < 2e-16 ***
## FEWTEETH 0.49006 0.02158 22.710 < 2e-16 ***
## MOSTTEETH 0.20118 0.01941 10.363 < 2e-16 ***
## MALE 0.77585 0.01503 51.605 < 2e-16 ***
## BLACK -0.30651 0.02853 -10.744 < 2e-16 ***
## HISPANIC -0.23566 0.03543 -6.651 2.90e-11 ***
## ASIAN -0.34785 0.07741 -4.493 7.01e-06 ***
## OTHRACE 0.25383 0.03113 8.153 3.55e-16 ***
## AGE1 0.39809 0.02163 18.407 < 2e-16 ***
## AGE2 0.80590 0.02227 36.185 < 2e-16 ***
## AGE3 1.16883 0.02541 45.998 < 2e-16 ***
## LOWED 0.15413 0.02395 6.435 1.24e-10 ***
## COLLEGE -0.14838 0.01773 -8.369 < 2e-16 ***
## INC1 0.45387 0.02472 18.358 < 2e-16 ***
## INC2 0.30198 0.02082 14.503 < 2e-16 ***
## INC3 0.13680 0.02502 5.467 4.57e-08 ***
## INC4 0.10846 0.02327 4.660 3.16e-06 ***
## NODENVST 0.21551 0.01632 13.202 < 2e-16 ***
## NODIABETE -0.70883 0.01594 -44.470 < 2e-16 ***
## FRMRSMK 0.35071 0.01616 21.700 < 2e-16 ***
## SMOKE 0.51367 0.02246 22.873 < 2e-16 ***
## NOEXER 0.22483 0.01548 14.523 < 2e-16 ***
## UNDWT 0.25641 0.05710 4.490 7.11e-06 ***
## OVWT 0.08429 0.01845 4.569 4.90e-06 ***
## OBESE 0.20787 0.01913 10.864 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 156763 on 266108 degrees of freedom
## Residual deviance: 140691 on 266083 degrees of freedom
## AIC: 140743
##
## Number of Fisher Scoring iterations: 6
##
##
## > logFullModel <- glm(CVDINFR5 ~ NOTEETH + FEWTEETH +
## + MOSTTEETH + MALE + BLACK + HISPANIC + ASIAN + OTHRACE + AGE1 +
## + AGE2 + AGE3 + LOWED .... [TRUNCATED]
##
## > summary(logFullModel)
##
## Call:
## glm(formula = CVDINFR5 ~ NOTEETH + FEWTEETH + MOSTTEETH + MALE +
## BLACK + HISPANIC + ASIAN + OTHRACE + AGE1 + AGE2 + AGE3 +
## LOWED + SOMECOLL + COLLEGE + INC1 + INC2 + INC3 + INC4 +
## NODENVST + NODIABETE + FRMRSMK + SMOKE + NOEXER + UNDWT +
## OVWT + OBESE, family = "binomial", data = BRFSS)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -1.4537 -0.4532 -0.3282 -0.2347 3.0642
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -3.57592 0.03386 -105.615 < 2e-16 ***
## NOTEETH 0.62128 0.02476 25.090 < 2e-16 ***
## FEWTEETH 0.49110 0.02160 22.740 < 2e-16 ***
## MOSTTEETH 0.20154 0.01942 10.380 < 2e-16 ***
## MALE 0.77617 0.01504 51.618 < 2e-16 ***
## BLACK -0.30616 0.02853 -10.731 < 2e-16 ***
## HISPANIC -0.23536 0.03543 -6.643 3.08e-11 ***
## ASIAN -0.34816 0.07742 -4.497 6.88e-06 ***
## OTHRACE 0.25326 0.03114 8.134 4.17e-16 ***
## AGE1 0.39748 0.02163 18.373 < 2e-16 ***
## AGE2 0.80548 0.02227 36.162 < 2e-16 ***
## AGE3 1.16911 0.02541 46.007 < 2e-16 ***
## LOWED 0.16342 0.02517 6.492 8.46e-11 ***
## SOMECOLL 0.02251 0.01865 1.207 0.227
## COLLEGE -0.13705 0.02008 -6.825 8.79e-12 ***
## INC1 0.45549 0.02476 18.396 < 2e-16 ***
## INC2 0.30338 0.02085 14.547 < 2e-16 ***
## INC3 0.13739 0.02503 5.490 4.03e-08 ***
## INC4 0.10841 0.02327 4.658 3.20e-06 ***
## NODENVST 0.21640 0.01634 13.243 < 2e-16 ***
## NODIABETE -0.70871 0.01594 -44.462 < 2e-16 ***
## FRMRSMK 0.35038 0.01616 21.676 < 2e-16 ***
## SMOKE 0.51351 0.02246 22.866 < 2e-16 ***
## NOEXER 0.22572 0.01550 14.563 < 2e-16 ***
## UNDWT 0.25637 0.05710 4.490 7.14e-06 ***
## OVWT 0.08429 0.01845 4.569 4.90e-06 ***
## OBESE 0.20773 0.01913 10.856 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 156763 on 266108 degrees of freedom
## Residual deviance: 140689 on 266082 degrees of freedom
## AIC: 140743
##
## Number of Fisher Scoring iterations: 6
##
##
## > summary(logModel1)$aic
## [1] 151296.6
##
## > summary(logModel2)$aic
## [1] 146072.8
##
## > summary(logFullModel)$aic
## [1] 140743.2
##
## > FinalLogisticRegressionModel <- logFullModel
##
## > summary(FinalLogisticRegressionModel)
##
## Call:
## glm(formula = CVDINFR5 ~ NOTEETH + FEWTEETH + MOSTTEETH + MALE +
## BLACK + HISPANIC + ASIAN + OTHRACE + AGE1 + AGE2 + AGE3 +
## LOWED + SOMECOLL + COLLEGE + INC1 + INC2 + INC3 + INC4 +
## NODENVST + NODIABETE + FRMRSMK + SMOKE + NOEXER + UNDWT +
## OVWT + OBESE, family = "binomial", data = BRFSS)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -1.4537 -0.4532 -0.3282 -0.2347 3.0642
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -3.57592 0.03386 -105.615 < 2e-16 ***
## NOTEETH 0.62128 0.02476 25.090 < 2e-16 ***
## FEWTEETH 0.49110 0.02160 22.740 < 2e-16 ***
## MOSTTEETH 0.20154 0.01942 10.380 < 2e-16 ***
## MALE 0.77617 0.01504 51.618 < 2e-16 ***
## BLACK -0.30616 0.02853 -10.731 < 2e-16 ***
## HISPANIC -0.23536 0.03543 -6.643 3.08e-11 ***
## ASIAN -0.34816 0.07742 -4.497 6.88e-06 ***
## OTHRACE 0.25326 0.03114 8.134 4.17e-16 ***
## AGE1 0.39748 0.02163 18.373 < 2e-16 ***
## AGE2 0.80548 0.02227 36.162 < 2e-16 ***
## AGE3 1.16911 0.02541 46.007 < 2e-16 ***
## LOWED 0.16342 0.02517 6.492 8.46e-11 ***
## SOMECOLL 0.02251 0.01865 1.207 0.227
## COLLEGE -0.13705 0.02008 -6.825 8.79e-12 ***
## INC1 0.45549 0.02476 18.396 < 2e-16 ***
## INC2 0.30338 0.02085 14.547 < 2e-16 ***
## INC3 0.13739 0.02503 5.490 4.03e-08 ***
## INC4 0.10841 0.02327 4.658 3.20e-06 ***
## NODENVST 0.21640 0.01634 13.243 < 2e-16 ***
## NODIABETE -0.70871 0.01594 -44.462 < 2e-16 ***
## FRMRSMK 0.35038 0.01616 21.676 < 2e-16 ***
## SMOKE 0.51351 0.02246 22.866 < 2e-16 ***
## NOEXER 0.22572 0.01550 14.563 < 2e-16 ***
## UNDWT 0.25637 0.05710 4.490 7.14e-06 ***
## OVWT 0.08429 0.01845 4.569 4.90e-06 ***
## OBESE 0.20773 0.01913 10.856 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 156763 on 266108 degrees of freedom
## Residual deviance: 140689 on 266082 degrees of freedom
## AIC: 140743
##
## Number of Fisher Scoring iterations: 6
##
##
## > Tidy_LogModel_a <- tidy(FinalLogisticRegressionModel)
##
## > Tidy_LogFinal <- subset(Tidy_LogModel_a, term != "(Intercept)")
##
## > Tidy_LogFinal$OR <- exp(Tidy_LogFinal$estimate)
##
## > Tidy_LogFinal$LL <- exp(Tidy_LogFinal$estimate - (1.96 *
## + Tidy_LogFinal$std.error))
##
## > Tidy_LogFinal$UL <- exp(Tidy_LogFinal$estimate + (1.96 *
## + Tidy_LogFinal$std.error))
##
## > write.csv(Tidy_LogFinal, file = "../output/logistic/LogisticRegressionModel.csv")
##
## > p1 <- ggplot(Tidy_LogFinal, aes(x = term, y = OR,
## + ymin = LL, ymax = UL)) + geom_pointrange(aes(col = factor(term)),
## + position = positio .... [TRUNCATED]
##
## > ggsave("../output/figures/LogModelOR.png", p1)
## Saving 7 x 5 in image
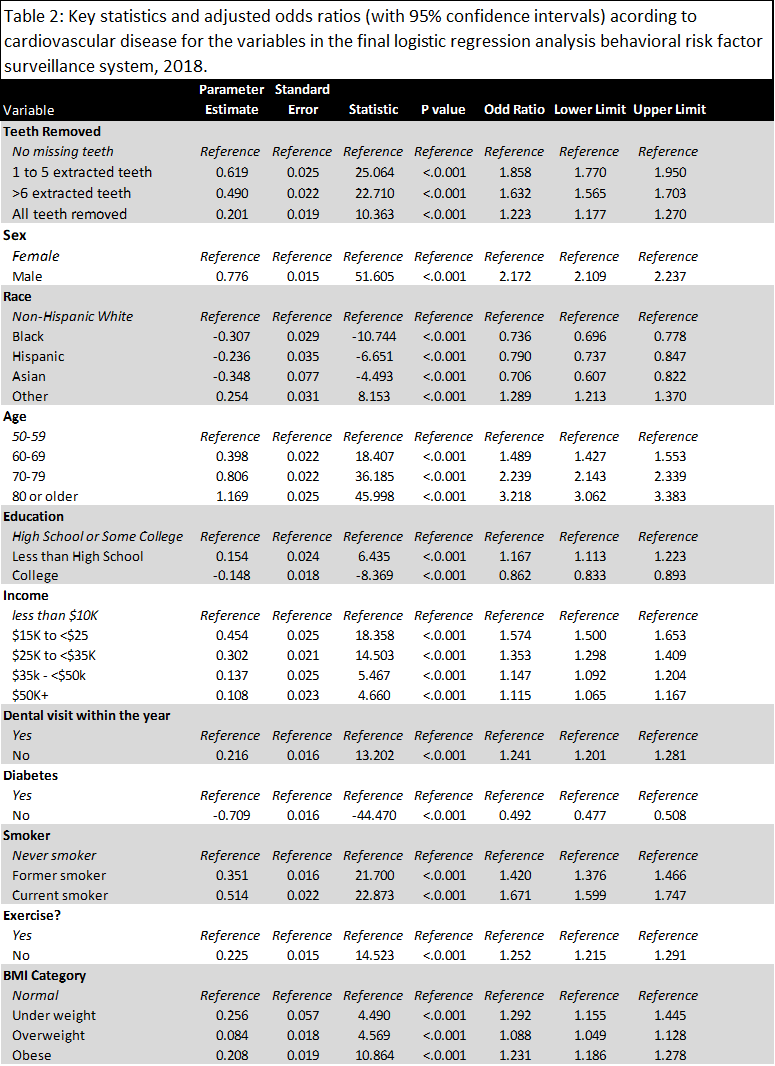
Full Logistical Model Parameters (with Odds Ratios and Limits)
Full Logistic Model Odds Ratio Plot
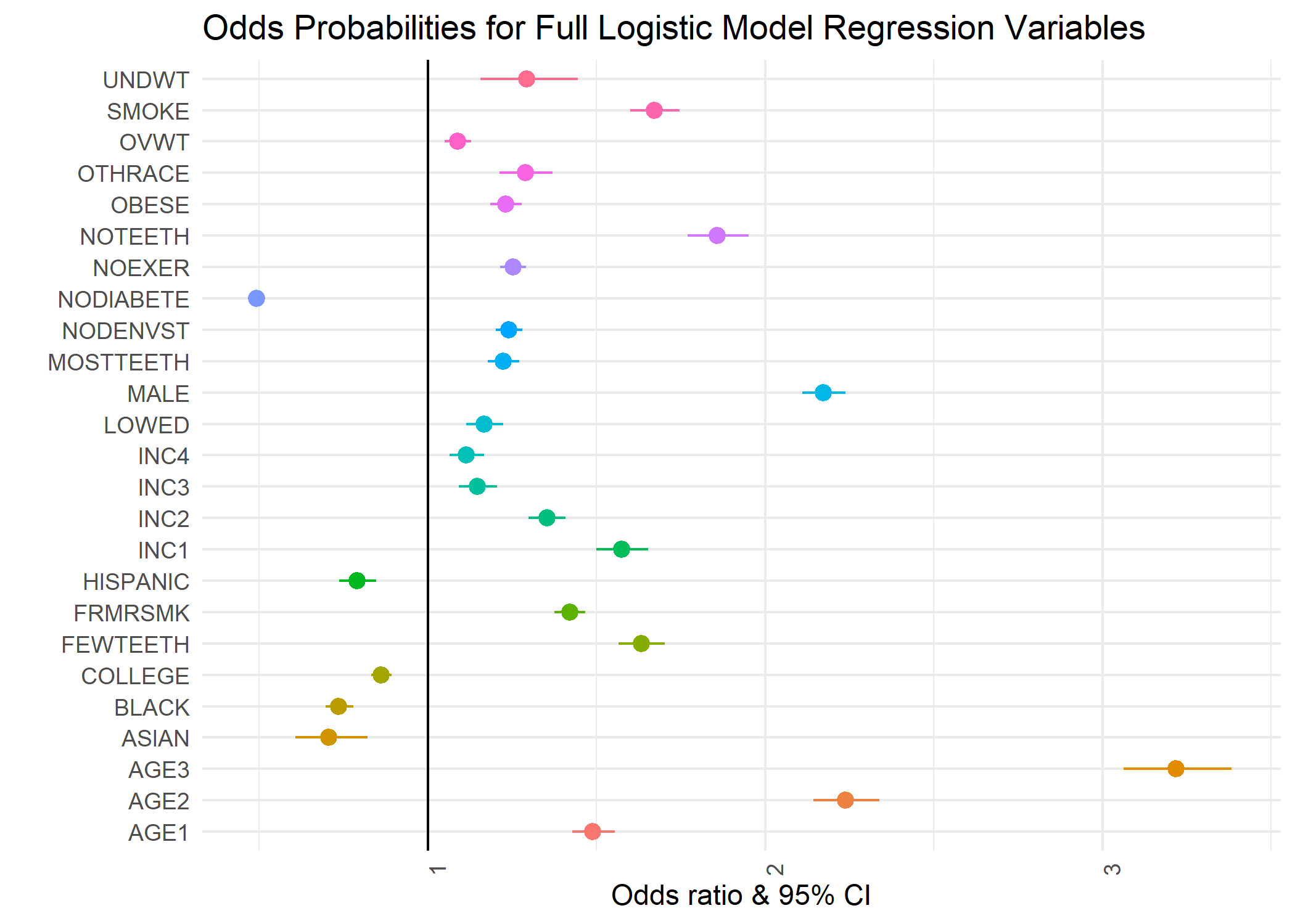
This concluded the logistic regression modeling.
Return to the Main Overview