Preparing the Data
First created on Nov 7, 2019. Updated on Nov 09, 2019
- Step 1: Read in the BRFSS 2018 survey data set and only keep key variables
- Step 2: Re-code and create dummy variables
- Step 3: Data reduction
- Step 4: Write out final data set for further analyses
Step 1: Read in the BRFSS 2018 survey data set and only keep key variables
source("../code/100_Dependencies.R", echo = TRUE)
##
## > library(foreign)
##
## > library(Hmisc)
## Loading required package: lattice
## Loading required package: survival
## Loading required package: Formula
## Loading required package: ggplot2
##
## Attaching package: 'Hmisc'
## The following objects are masked from 'package:base':
##
## format.pval, units
##
## > library(e1071)
##
## Attaching package: 'e1071'
## The following object is masked from 'package:Hmisc':
##
## impute
##
## > library(gtools)
##
## Attaching package: 'gtools'
## The following object is masked from 'package:e1071':
##
## permutations
##
## > library(MASS)
##
## > library(ggplot2)
##
## > library(gridExtra)
##
## > library(broom)
##
## > library(scales)
##
## > library(ggfortify)
##
## > library(gvlma)
source("../code/105_Read in BRFSS XPT.R", echo = TRUE)
##
## > BRFSS_a <- read.xport("../../large_data_sets/LLCP2018.xpt")
##
## > colnames(BRFSS_a)
## [1] "X_STATE" "FMONTH" "IDATE" "IMONTH" "IDAY"
## [6] "IYEAR" "DISPCODE" "SEQNO" "X_PSU" "CTELENM1"
## [11] "PVTRESD1" "COLGHOUS" "STATERE1" "CELLFON4" "LADULT"
## [16] "NUMADULT" "NUMMEN" "NUMWOMEN" "SAFETIME" "CTELNUM1"
## [21] "CELLFON5" "CADULT" "PVTRESD3" "CCLGHOUS" "CSTATE1"
## [26] "LANDLINE" "HHADULT" "GENHLTH" "PHYSHLTH" "MENTHLTH"
## [31] "POORHLTH" "HLTHPLN1" "PERSDOC2" "MEDCOST" "CHECKUP1"
## [36] "EXERANY2" "SLEPTIM1" "CVDINFR4" "CVDCRHD4" "CVDSTRK3"
## [41] "ASTHMA3" "ASTHNOW" "CHCSCNCR" "CHCOCNCR" "CHCCOPD1"
## [46] "HAVARTH3" "ADDEPEV2" "CHCKDNY1" "DIABETE3" "DIABAGE2"
## [51] "LASTDEN4" "RMVTETH4" "SEX1" "MARITAL" "EDUCA"
## [56] "RENTHOM1" "NUMHHOL3" "NUMPHON3" "CPDEMO1B" "VETERAN3"
## [61] "EMPLOY1" "CHILDREN" "INCOME2" "WEIGHT2" "HEIGHT3"
## [66] "PREGNANT" "DEAF" "BLIND" "DECIDE" "DIFFWALK"
## [71] "DIFFDRES" "DIFFALON" "SMOKE100" "SMOKDAY2" "STOPSMK2"
## [76] "LASTSMK2" "USENOW3" "ALCDAY5" "AVEDRNK2" "DRNK3GE5"
## [81] "MAXDRNKS" "FLUSHOT6" "FLSHTMY2" "IMFVPLAC" "PNEUVAC4"
## [86] "FALL12MN" "FALLINJ3" "SEATBELT" "DRNKDRI2" "HADMAM"
## [91] "HOWLONG" "HADPAP2" "LASTPAP2" "HPVTEST" "HPLSTTST"
## [96] "HADHYST2" "PCPSAAD3" "PCPSADI1" "PCPSARE1" "PSATEST1"
## [101] "PSATIME" "PCPSARS1" "BLDSTOOL" "LSTBLDS3" "HADSIGM3"
## [106] "HADSGCO1" "LASTSIG3" "HIVTST6" "HIVTSTD3" "HIVRISK5"
## [111] "PDIABTST" "PREDIAB1" "INSULIN" "BLDSUGAR" "FEETCHK3"
## [116] "DOCTDIAB" "CHKHEMO3" "FEETCHK" "EYEEXAM1" "DIABEYE"
## [121] "DIABEDU" "MEDICARE" "HLTHCVR1" "DELAYME1" "DLYOTHER"
## [126] "NOCOV121" "LSTCOVRG" "DRVISITS" "MEDSCOS1" "CARERCVD"
## [131] "MEDBILL1" "CIMEMLOS" "CDHOUSE" "CDASSIST" "CDHELP"
## [136] "CDSOCIAL" "CDDISCUS" "CAREGIV1" "CRGVREL2" "CRGVLNG1"
## [141] "CRGVHRS1" "CRGVPRB2" "CRGVPERS" "CRGVHOUS" "CRGVMST3"
## [146] "CRGVEXPT" "ECIGARET" "ECIGNOW" "MARIJAN1" "USEMRJN2"
## [151] "RSNMRJN1" "ADSLEEP" "SLEPDAY1" "SLEPSNO2" "SLEPBRTH"
## [156] "ADPLEAS1" "ADDOWN1" "FEELNERV" "STOPWORY" "COPDCOGH"
## [161] "COPDFLEM" "COPDBRTH" "COPDBTST" "COPDSMOK" "INDORTAN"
## [166] "NUMBURN3" "SUNPRTCT" "WKDAYOUT" "WKENDOUT" "LCSFIRST"
## [171] "LCSLAST" "LCSNUMCG" "LCSCTSCN" "CNCRDIFF" "CNCRAGE"
## [176] "CNCRTYP1" "CSRVTRT2" "CSRVDOC1" "CSRVSUM" "CSRVRTRN"
## [181] "CSRVINST" "CSRVINSR" "CSRVDEIN" "CSRVCLIN" "CSRVPAIN"
## [186] "CSRVCTL1" "PCPSADE1" "PCDMDEC1" "PROFEXAM" "LENGEXAM"
## [191] "HPVADVC2" "HPVADSHT" "TETANUS1" "SHINGLE2" "SOMALE"
## [196] "SOFEMALE" "TRNSGNDR" "RCSGENDR" "RCSRLTN2" "CASTHDX2"
## [201] "CASTHNO2" "QSTVER" "QSTLANG" "X_METSTAT" "X_URBSTAT"
## [206] "MSCODE" "X_STSTR" "X_STRWT" "X_RAWRAKE" "X_WT2RAKE"
## [211] "X_IMPRACE" "X_CHISPNC" "X_CRACE1" "X_CPRACE" "X_CLLCPWT"
## [216] "X_DUALUSE" "X_DUALCOR" "X_LLCPWT2" "X_LLCPWT" "X_RFHLTH"
## [221] "X_PHYS14D" "X_MENT14D" "X_HCVU651" "X_TOTINDA" "X_MICHD"
## [226] "X_LTASTH1" "X_CASTHM1" "X_ASTHMS1" "X_DRDXAR1" "X_EXTETH3"
## [231] "X_ALTETH3" "X_DENVST3" "X_PRACE1" "X_MRACE1" "X_HISPANC"
## [236] "X_RACE" "X_RACEG21" "X_RACEGR3" "X_RACE_G1" "X_AGEG5YR"
## [241] "X_AGE65YR" "X_AGE80" "X_AGE_G" "HTIN4" "HTM4"
## [246] "WTKG3" "X_BMI5" "X_BMI5CAT" "X_RFBMI5" "X_CHLDCNT"
## [251] "X_EDUCAG" "X_INCOMG" "X_SMOKER3" "X_RFSMOK3" "DRNKANY5"
## [256] "DROCDY3_" "X_RFBING5" "X_DRNKWEK" "X_RFDRHV6" "X_FLSHOT6"
## [261] "X_PNEUMO3" "X_RFSEAT2" "X_RFSEAT3" "X_DRNKDRV" "X_RFMAM21"
## [266] "X_MAM5022" "X_RFPAP34" "X_RFPSA22" "X_RFBLDS3" "X_COL10YR"
## [271] "X_HFOB3YR" "X_FS5YR" "X_FOBTFS" "X_CRCREC" "X_AIDTST3"
##
## > nrow(BRFSS_a)
## [1] 437436
source("../code/110_Keep vars.R", echo = TRUE)
##
## > BRFSS_b <- BRFSS_a
##
## > BRFSSVarList <- c("CVDINFR4", "RMVTETH4", "SEX1",
## + "X_HISPANC", "X_MRACE1", "X_AGEG5YR", "EDUCA", "INCOME2",
## + "X_DENVST3", "DIABETE3", " ..." ... [TRUNCATED]
##
## > BRFSS_b <- BRFSS_b[BRFSSVarList]
##
## > colnames(BRFSS_b)
## [1] "CVDINFR4" "RMVTETH4" "SEX1" "X_HISPANC" "X_MRACE1"
## [6] "X_AGEG5YR" "EDUCA" "INCOME2" "X_DENVST3" "DIABETE3"
## [11] "X_SMOKER3" "EXERANY2" "X_BMI5CAT"
##
## > nrow(BRFSS_b)
## [1] 437436
Step 2: Re-code and create dummy variables
The next step in the process is to recode the variables from the BRFSS 2018 survey to better suit these analyses, and to create some additional dummy (indicator) variables for use in a linear regression analysis. Dummy variables are useful because they enable us to use a single regression equation to represent multiple groups.
While the BRFSS data set used here is relatively clean, some of the additional recoding allowed for some additional cleaning. Particularly, don’t know/refused/missing data was consistently recoded.
source("../code/115_Make outcome exposure vars.R", echo = TRUE)
##
## > BRFSS_c <- BRFSS_b
##
## > BRFSS_c$CVDINFR5 <- 9
##
## > BRFSS_c$CVDINFR5[BRFSS_c$CVDINFR4 == 1] <- 1
##
## > BRFSS_c$CVDINFR5[BRFSS_c$CVDINFR4 == 2] <- 0
##
## > table(BRFSS_c$CVDINFR4, BRFSS_c$CVDINFR5)
##
## 0 1 9
## 1 0 26412 0
## 2 408706 0 0
## 7 0 0 2055
## 9 0 0 240
##
## > BRFSS_c$RMVTETH5 <- 9
##
## > BRFSS_c$RMVTETH5[BRFSS_c$RMVTETH4 == 8] <- 0
##
## > BRFSS_c$RMVTETH5[BRFSS_c$RMVTETH4 == 1] <- 1
##
## > BRFSS_c$RMVTETH5[BRFSS_c$RMVTETH4 == 2] <- 2
##
## > BRFSS_c$RMVTETH5[BRFSS_c$RMVTETH4 == 3] <- 3
##
## > table(BRFSS_c$RMVTETH4, BRFSS_c$RMVTETH5)
##
## 0 1 2 3 9
## 1 0 127234 0 0 0
## 2 0 0 51635 0 0
## 3 0 0 0 29430 0
## 7 0 0 0 0 9687
## 8 218715 0 0 0 0
## 9 0 0 0 0 707
##
## > BRFSS_c$MOSTTEETH <- 0
##
## > BRFSS_c$FEWTEETH <- 0
##
## > BRFSS_c$NOTEETH <- 0
##
## > BRFSS_c$MOSTTEETH[BRFSS_c$RMVTETH5 == 1] <- 1
##
## > BRFSS_c$FEWTEETH[BRFSS_c$RMVTETH5 == 2] <- 1
##
## > BRFSS_c$NOTEETH[BRFSS_c$RMVTETH5 == 3] <- 1
##
## > table(BRFSS_c$RMVTETH5, BRFSS_c$MOSTTEETH)
##
## 0 1
## 0 218715 0
## 1 0 127234
## 2 51635 0
## 3 29430 0
## 9 10422 0
##
## > table(BRFSS_c$RMVTETH5, BRFSS_c$FEWTEETH)
##
## 0 1
## 0 218715 0
## 1 127234 0
## 2 0 51635
## 3 29430 0
## 9 10422 0
##
## > table(BRFSS_c$RMVTETH5, BRFSS_c$NOTEETH)
##
## 0 1
## 0 218715 0
## 1 127234 0
## 2 51635 0
## 3 0 29430
## 9 10422 0
source("../code/120_Make sex race vars.R", echo = TRUE)
##
## > BRFSS_c$SEX2 <- 9
##
## > BRFSS_c$SEX2[BRFSS_c$SEX1 == 1] <- 1
##
## > BRFSS_c$SEX2[BRFSS_c$SEX1 == 2] <- 0
##
## > table(BRFSS_c$SEX1, BRFSS_c$SEX2)
##
## 0 1 9
## 1 0 197412 0
## 2 238911 0 0
## 7 0 0 431
## 9 0 0 682
##
## > BRFSS_c$MALE <- 0
##
## > BRFSS_c$MALE[BRFSS_c$SEX2 == 1] <- 1
##
## > table(BRFSS_c$SEX2, BRFSS_c$MALE)
##
## 0 1
## 0 238911 0
## 1 0 197412
## 9 1113 0
##
## > BRFSS_c$HISPANC2 <- 9
##
## > BRFSS_c$HISPANC2[BRFSS_c$X_HISPANC == 1] <- 1
##
## > BRFSS_c$HISPANC2[BRFSS_c$X_HISPANC == 2] <- 0
##
## > table(BRFSS_c$X_HISPANC, BRFSS_c$HISPANC2)
##
## 0 1 9
## 1 0 36941 0
## 2 395932 0 0
## 9 0 0 4563
##
## > BRFSS_c$HISPANIC <- 0
##
## > BRFSS_c$HISPANIC[BRFSS_c$HISPANC2 == 1] <- 1
##
## > table(BRFSS_c$HISPANC2, BRFSS_c$HISPANIC)
##
## 0 1
## 0 395932 0
## 1 0 36941
## 9 4563 0
##
## > BRFSS_c$RACEGRP <- 9
##
## > BRFSS_c$RACEGRP[BRFSS_c$X_MRACE1 == 1 & BRFSS_c$HISPANIC ==
## + 0] <- 0
##
## > BRFSS_c$RACEGRP[BRFSS_c$X_MRACE1 == 2 & BRFSS_c$HISPANIC ==
## + 0] <- 1
##
## > BRFSS_c$RACEGRP[BRFSS_c$X_MRACE1 == 3 & BRFSS_c$HISPANIC ==
## + 0] <- 4
##
## > BRFSS_c$RACEGRP[BRFSS_c$X_MRACE1 == 4 & BRFSS_c$HISPANIC ==
## + 0] <- 3
##
## > BRFSS_c$RACEGRP[BRFSS_c$X_MRACE1 == 5 & BRFSS_c$HISPANIC ==
## + 0] <- 4
##
## > BRFSS_c$RACEGRP[BRFSS_c$X_MRACE1 == 6 & BRFSS_c$HISPANIC ==
## + 0] <- 4
##
## > BRFSS_c$RACEGRP[BRFSS_c$X_MRACE1 == 7 & BRFSS_c$HISPANIC ==
## + 0] <- 4
##
## > BRFSS_c$RACEGRP[BRFSS_c$HISPANIC == 1] <- 2
##
## > table(BRFSS_b$X_MRACE1, BRFSS_c$RACEGRP)
##
## 0 1 2 3 4 9
## 1 326148 0 19563 0 0 0
## 2 0 36179 1683 0 0 0
## 3 0 0 1444 0 7687 0
## 4 0 0 264 9994 0 0
## 5 0 0 486 0 2131 0
## 6 0 0 8192 0 3449 0
## 7 0 0 977 0 8595 0
## 77 0 0 3318 0 0 935
## 99 0 0 1014 0 0 5345
##
## > BRFSS_c$BLACK <- 0
##
## > BRFSS_c$ASIAN <- 0
##
## > BRFSS_c$OTHRACE <- 0
##
## > BRFSS_c$BLACK[BRFSS_c$RACEGRP == 1] <- 1
##
## > BRFSS_c$ASIAN[BRFSS_c$RACEGRP == 3] <- 1
##
## > BRFSS_c$OTHRACE[BRFSS_c$RACEGRP == 4] <- 1
##
## > table(BRFSS_c$RACEGRP, BRFSS_c$BLACK)
##
## 0 1
## 0 326148 0
## 1 0 36179
## 2 36941 0
## 3 9994 0
## 4 21862 0
## 9 6312 0
##
## > table(BRFSS_c$RACEGRP, BRFSS_c$ASIAN)
##
## 0 1
## 0 326148 0
## 1 36179 0
## 2 36941 0
## 3 0 9994
## 4 21862 0
## 9 6312 0
##
## > table(BRFSS_c$RACEGRP, BRFSS_c$OTHRACE)
##
## 0 1
## 0 326148 0
## 1 36179 0
## 2 36941 0
## 3 9994 0
## 4 0 21862
## 9 6312 0
source("../code/125_Make age educ income vars.R", echo = TRUE)
##
## > BRFSS_c$AGEG5YR2 <- 9
##
## > BRFSS_c$AGEG5YR2[BRFSS_c$X_AGEG5YR > 6 & BRFSS_c$X_AGEG5YR <
## + 9] <- 0
##
## > BRFSS_c$AGEG5YR2[BRFSS_c$X_AGEG5YR > 8 & BRFSS_c$X_AGEG5YR <
## + 11] <- 1
##
## > BRFSS_c$AGEG5YR2[BRFSS_c$X_AGEG5YR > 10 & BRFSS_c$X_AGEG5YR <
## + 13] <- 2
##
## > BRFSS_c$AGEG5YR2[BRFSS_c$X_AGEG5YR == 13] <- 3
##
## > table(BRFSS_c$X_AGEG5YR, BRFSS_c$AGEG5YR2)
##
## 0 1 2 3 9
## 1 0 0 0 0 26005
## 2 0 0 0 0 22286
## 3 0 0 0 0 24269
## 4 0 0 0 0 26118
## 5 0 0 0 0 25583
## 6 0 0 0 0 28846
## 7 35060 0 0 0 0
## 8 42160 0 0 0 0
## 9 0 46994 0 0 0
## 10 0 47329 0 0 0
## 11 0 0 40930 0 0
## 12 0 0 28668 0 0
## 13 0 0 0 34716 0
## 14 0 0 0 0 8472
##
## > BRFSS_c$AGE1 <- 0
##
## > BRFSS_c$AGE2 <- 0
##
## > BRFSS_c$AGE3 <- 0
##
## > BRFSS_c$AGE1[BRFSS_c$AGEG5YR2 == 1] <- 1
##
## > BRFSS_c$AGE2[BRFSS_c$AGEG5YR2 == 2] <- 1
##
## > BRFSS_c$AGE3[BRFSS_c$AGEG5YR2 == 3] <- 1
##
## > table(BRFSS_c$AGEG5YR2, BRFSS_c$AGE1)
##
## 0 1
## 0 77220 0
## 1 0 94323
## 2 69598 0
## 3 34716 0
## 9 161579 0
##
## > table(BRFSS_c$AGEG5YR2, BRFSS_c$AGE2)
##
## 0 1
## 0 77220 0
## 1 94323 0
## 2 0 69598
## 3 34716 0
## 9 161579 0
##
## > table(BRFSS_c$AGEG5YR2, BRFSS_c$AGE3)
##
## 0 1
## 0 77220 0
## 1 94323 0
## 2 69598 0
## 3 0 34716
## 9 161579 0
##
## > BRFSS_c$EDGRP <- 9
##
## > BRFSS_c$EDGRP[BRFSS_c$EDUCA < 4] <- 0
##
## > BRFSS_c$EDGRP[BRFSS_c$EDUCA == 4] <- 1
##
## > BRFSS_c$EDGRP[BRFSS_c$EDUCA == 5] <- 2
##
## > BRFSS_c$EDGRP[BRFSS_c$EDUCA == 6] <- 3
##
## > table(BRFSS_b$EDUCA, BRFSS_c$EDGRP)
##
## 0 1 2 3 9
## 1 666 0 0 0 0
## 2 10446 0 0 0 0
## 3 21456 0 0 0 0
## 4 0 119038 0 0 0
## 5 0 0 119980 0 0
## 6 0 0 0 164229 0
## 9 0 0 0 0 1587
##
## > BRFSS_c$LOWED <- 0
##
## > BRFSS_c$SOMECOLL <- 0
##
## > BRFSS_c$COLLEGE <- 0
##
## > BRFSS_c$LOWED[BRFSS_c$EDGRP == 0] <- 1
##
## > BRFSS_c$SOMECOLL[BRFSS_c$EDGRP == 2] <- 1
##
## > BRFSS_c$COLLEGE[BRFSS_c$EDGRP == 3] <- 1
##
## > table(BRFSS_c$EDGRP, BRFSS_c$LOWED)
##
## 0 1
## 0 0 32568
## 1 119038 0
## 2 119980 0
## 3 164229 0
## 9 1621 0
##
## > table(BRFSS_c$EDGRP, BRFSS_c$SOMECOLL)
##
## 0 1
## 0 32568 0
## 1 119038 0
## 2 0 119980
## 3 164229 0
## 9 1621 0
##
## > table(BRFSS_c$EDGRP, BRFSS_c$COLLEGE)
##
## 0 1
## 0 32568 0
## 1 119038 0
## 2 119980 0
## 3 0 164229
## 9 1621 0
##
## > BRFSS_c$INCOME3 <- 9
##
## > BRFSS_c$INCOME3[BRFSS_c$INCOME2 < 3] <- 0
##
## > BRFSS_c$INCOME3[BRFSS_c$INCOME2 > 2 & BRFSS_c$INCOME2 <
## + 5] <- 1
##
## > BRFSS_c$INCOME3[BRFSS_c$INCOME2 > 4 & BRFSS_c$INCOME2 <
## + 6] <- 2
##
## > BRFSS_c$INCOME3[BRFSS_c$INCOME2 > 5 & BRFSS_c$INCOME2 <
## + 7] <- 3
##
## > BRFSS_c$INCOME3[BRFSS_c$INCOME2 > 6 & BRFSS_c$INCOME2 <
## + 9] <- 4
##
## > table(BRFSS_c$INCOME2, BRFSS_c$INCOME3)
##
## 0 1 2 3 4 9
## 1 16762 0 0 0 0 0
## 2 18034 0 0 0 0 0
## 3 0 25850 0 0 0 0
## 4 0 32218 0 0 0 0
## 5 0 0 37830 0 0 0
## 6 0 0 0 49558 0 0
## 7 0 0 0 0 58098 0
## 8 0 0 0 0 122968 0
## 77 0 0 0 0 0 33279
## 99 0 0 0 0 0 38445
##
## > BRFSS_c$INC1 <- 0
##
## > BRFSS_c$INC2 <- 0
##
## > BRFSS_c$INC3 <- 0
##
## > BRFSS_c$INC4 <- 0
##
## > BRFSS_c$INC1[BRFSS_c$INCOME3 == 0] <- 1
##
## > BRFSS_c$INC2[BRFSS_c$INCOME3 == 1] <- 1
##
## > BRFSS_c$INC3[BRFSS_c$INCOME3 == 2] <- 1
##
## > BRFSS_c$INC4[BRFSS_c$INCOME3 == 3] <- 1
##
## > table(BRFSS_c$INCOME3, BRFSS_c$INC1)
##
## 0 1
## 0 0 34796
## 1 58068 0
## 2 37830 0
## 3 49558 0
## 4 181066 0
## 9 76118 0
##
## > table(BRFSS_c$INCOME3, BRFSS_c$INC2)
##
## 0 1
## 0 34796 0
## 1 0 58068
## 2 37830 0
## 3 49558 0
## 4 181066 0
## 9 76118 0
##
## > table(BRFSS_c$INCOME3, BRFSS_c$INC3)
##
## 0 1
## 0 34796 0
## 1 58068 0
## 2 0 37830
## 3 49558 0
## 4 181066 0
## 9 76118 0
##
## > table(BRFSS_c$INCOME3, BRFSS_c$INC4)
##
## 0 1
## 0 34796 0
## 1 58068 0
## 2 37830 0
## 3 0 49558
## 4 181066 0
## 9 76118 0
source("../code/130_Make dent diabetes smoker vars.R", echo = TRUE)
##
## > BRFSS_c$DENVST4 <- 9
##
## > BRFSS_c$DENVST4[BRFSS_c$X_DENVST3 == 1] <- 0
##
## > BRFSS_c$DENVST4[BRFSS_c$X_DENVST3 == 2] <- 1
##
## > table(BRFSS_c$X_DENVST3, BRFSS_c$DENVST4)
##
## 0 1 9
## 1 296047 0 0
## 2 0 136629 0
## 9 0 0 4760
##
## > BRFSS_c$NODENVST <- 0
##
## > BRFSS_c$NODENVST[BRFSS_c$DENVST4 == 1] <- 1
##
## > table(BRFSS_c$DENVST4, BRFSS_c$NODENVST)
##
## 0 1
## 0 296047 0
## 1 0 136629
## 9 4760 0
##
## > BRFSS_c$DIABETE4 <- 9
##
## > BRFSS_c$DIABETE4[BRFSS_c$DIABETE3 > 1 & BRFSS_c$DIABETE3 <
## + 7] <- 0
##
## > BRFSS_c$DIABETE4[BRFSS_c$DIABETE3 == 1] <- 1
##
## > table(BRFSS_b$DIABETE3, BRFSS_c$DIABETE4)
##
## 0 1 9
## 1 0 60703 0
## 2 3857 0 0
## 3 363757 0 0
## 4 8263 0 0
## 7 0 0 567
## 9 0 0 265
##
## > BRFSS_c$NODIABETE <- 0
##
## > BRFSS_c$NODIABETE[BRFSS_c$DIABETE4 == 0] <- 1
##
## > table(BRFSS_c$DIABETE4, BRFSS_c$NODIABETE)
##
## 0 1
## 0 0 375877
## 1 60703 0
## 9 856 0
##
## > BRFSS_c$SMOKER4 <- 9
##
## > BRFSS_c$SMOKER4[BRFSS_c$X_SMOKER3 == 4] <- 0
##
## > BRFSS_c$SMOKER4[BRFSS_c$X_SMOKER3 == 3] <- 1
##
## > BRFSS_c$SMOKER4[BRFSS_c$X_SMOKER3 < 3] <- 2
##
## > table(BRFSS_c$X_SMOKER3, BRFSS_c$SMOKER4)
##
## 0 1 2 9
## 1 0 0 43633 0
## 2 0 0 17639 0
## 3 0 118754 0 0
## 4 240594 0 0 0
## 9 0 0 0 16816
##
## > BRFSS_c$FRMRSMK <- 0
##
## > BRFSS_c$SMOKE <- 0
##
## > BRFSS_c$FRMRSMK[BRFSS_c$SMOKER4 == 1] <- 1
##
## > BRFSS_c$SMOKE[BRFSS_c$SMOKER4 == 2] <- 1
##
## > table(BRFSS_c$SMOKER4, BRFSS_c$FRMRSMK)
##
## 0 1
## 0 240594 0
## 1 0 118754
## 2 61272 0
## 9 16816 0
##
## > table(BRFSS_c$SMOKER4, BRFSS_c$SMOKE)
##
## 0 1
## 0 240594 0
## 1 118754 0
## 2 0 61272
## 9 16816 0
source("../code/135_Make exer bmi vars.R", echo = TRUE)
##
## > BRFSS_c$EXERANY3 <- 9
##
## > BRFSS_c$EXERANY3[BRFSS_c$EXERANY2 == 1] <- 1
##
## > BRFSS_c$EXERANY3[BRFSS_c$EXERANY2 == 2] <- 0
##
## > table(BRFSS_c$EXERANY2, BRFSS_c$EXERANY3)
##
## 0 1 9
## 1 0 326472 0
## 2 110269 0 0
## 7 0 0 482
## 9 0 0 188
##
## > BRFSS_c$NOEXER <- 0
##
## > BRFSS_c$NOEXER[BRFSS_c$EXERANY3 == 0] <- 1
##
## > table(BRFSS_c$EXERANY3, BRFSS_c$NOEXER)
##
## 0 1
## 0 0 110269
## 1 326472 0
## 9 695 0
##
## > BRFSS_c$BMICAT <- 9
##
## > BRFSS_c$BMICAT[BRFSS_c$X_BMI5CAT == 1] <- 0
##
## > BRFSS_c$BMICAT[BRFSS_c$X_BMI5CAT == 2] <- 1
##
## > BRFSS_c$BMICAT[BRFSS_c$X_BMI5CAT == 3] <- 2
##
## > BRFSS_c$BMICAT[BRFSS_c$X_BMI5CAT == 4] <- 3
##
## > table(BRFSS_c$X_BMI5CAT, BRFSS_c$BMICAT)
##
## 0 1 2 3 9
## 1 6776 0 0 0 0
## 2 0 123522 0 0 0
## 3 0 0 143878 0 0
## 4 0 0 0 127998 0
##
## > BRFSS_c$UNDWT <- 0
##
## > BRFSS_c$OVWT <- 0
##
## > BRFSS_c$OBESE <- 0
##
## > BRFSS_c$UNDWT[BRFSS_c$BMICAT == 0] <- 1
##
## > BRFSS_c$OVWT[BRFSS_c$BMICAT == 2] <- 1
##
## > BRFSS_c$OBESE[BRFSS_c$BMICAT == 3] <- 1
##
## > table(BRFSS_c$BMICAT, BRFSS_c$UNDWT)
##
## 0 1
## 0 0 6776
## 1 123522 0
## 2 143878 0
## 3 127998 0
## 9 35262 0
##
## > table(BRFSS_c$BMICAT, BRFSS_c$OVWT)
##
## 0 1
## 0 6776 0
## 1 123522 0
## 2 0 143878
## 3 127998 0
## 9 35262 0
##
## > table(BRFSS_c$BMICAT, BRFSS_c$OBESE)
##
## 0 1
## 0 6776 0
## 1 123522 0
## 2 143878 0
## 3 0 127998
## 9 35262 0
Step 3: Data reduction
Now that the data are clean and reorganize, data reduction is possible. The original BRFSS 2018 Survey data included data on 437436 participants and 275 variables.
Data reduction occured in 3 phases:
- Duplicating the authors of the original 2010 analysis, the sample population was limited to adults over 50 years of age. BRFSS 2018 data for participants with age<50 were removed.
- Records that did not include valid data on the dependent (outcome) variable were also excluded. BRFSS 2018 data without valid cardiovascular disease data in CVDINFR5 were removed.
- Records that did not include valid data on the independent (exposure) variable were excluded as well. BRFSS 2018 data without valid missing teeth data in RMVTETH5 were removed.
source("../code/175_Data reduction.R", echo = TRUE)
##
## > BRFSS_d <- subset(BRFSS_c, BRFSS_c$AGEG5YR2 != 9)
##
## > nrow(BRFSS_d)
## [1] 275857
##
## > BRFSS_e <- subset(BRFSS_d, BRFSS_d$CVDINFR5 != 9)
##
## > nrow(BRFSS_e)
## [1] 274309
##
## > BRFSS_f <- subset(BRFSS_e, BRFSS_e$RMVTETH5 != 9)
##
## > nrow(BRFSS_f)
## [1] 266109
Data reduction results
As stated above this inital BRFSS 2018 Survey data included records on 437436 participants and 275 variables. Each of the steps resulted in making the data set smaller
- Removing participants under 50 eliminated 161579 records.
- Removing participants with invalid dependent (outcome) variable data eliminated 1548 records.
- Removing participants with invalid independent (exposure) variable data eliminated 8200 records.
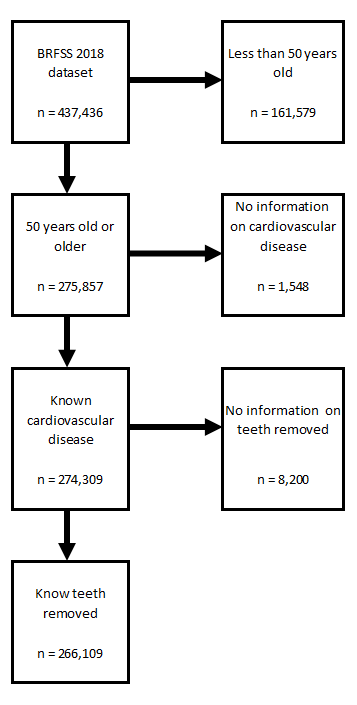
Step 4: Write out final data set for further analyses
The final data set included data on 274309 variables and 274309 participant. This final (and much smaller) data set was saved to speed up further analyses by requiring the loading of this data file.
source("../code/190_Write out data.R", echo = TRUE)
##
## > BRFSS <- BRFSS_f
##
## > write.csv(BRFSS, file = "../output/BRFSS.csv", row.names = FALSE)
##
## > str(BRFSS)
## 'data.frame': 266109 obs. of 52 variables:
## $ CVDINFR4 : num 2 2 2 2 2 2 2 2 2 2 ...
## $ RMVTETH4 : num 1 1 8 8 2 8 1 2 1 3 ...
## $ SEX1 : num 2 1 2 1 1 2 2 2 2 1 ...
## $ X_HISPANC: num 2 2 2 2 2 2 2 2 2 2 ...
## $ X_MRACE1 : num 1 1 1 1 1 1 1 2 1 1 ...
## $ X_AGEG5YR: num 13 10 12 8 13 8 11 12 10 11 ...
## $ EDUCA : num 6 4 6 4 4 5 4 3 3 5 ...
## $ INCOME2 : num 6 3 8 8 4 6 4 2 2 7 ...
## $ X_DENVST3: num 1 2 1 1 1 1 2 1 1 2 ...
## $ DIABETE3 : num 3 3 3 1 3 3 1 3 1 3 ...
## $ X_SMOKER3: num 4 4 4 3 3 4 4 4 4 4 ...
## $ EXERANY2 : num 2 1 1 2 2 1 2 2 1 1 ...
## $ X_BMI5CAT: num 2 3 2 3 3 4 4 4 4 4 ...
## $ CVDINFR5 : num 0 0 0 0 0 0 0 0 0 0 ...
## $ RMVTETH5 : num 1 1 0 0 2 0 1 2 1 3 ...
## $ MOSTTEETH: num 1 1 0 0 0 0 1 0 1 0 ...
## $ FEWTEETH : num 0 0 0 0 1 0 0 1 0 0 ...
## $ NOTEETH : num 0 0 0 0 0 0 0 0 0 1 ...
## $ SEX2 : num 0 1 0 1 1 0 0 0 0 1 ...
## $ MALE : num 0 1 0 1 1 0 0 0 0 1 ...
## $ HISPANC2 : num 0 0 0 0 0 0 0 0 0 0 ...
## $ HISPANIC : num 0 0 0 0 0 0 0 0 0 0 ...
## $ RACEGRP : num 0 0 0 0 0 0 0 1 0 0 ...
## $ BLACK : num 0 0 0 0 0 0 0 1 0 0 ...
## $ ASIAN : num 0 0 0 0 0 0 0 0 0 0 ...
## $ OTHRACE : num 0 0 0 0 0 0 0 0 0 0 ...
## $ AGEG5YR2 : num 3 1 2 0 3 0 2 2 1 2 ...
## $ AGE1 : num 0 1 0 0 0 0 0 0 1 0 ...
## $ AGE2 : num 0 0 1 0 0 0 1 1 0 1 ...
## $ AGE3 : num 1 0 0 0 1 0 0 0 0 0 ...
## $ EDGRP : num 3 1 3 1 1 2 1 0 0 2 ...
## $ LOWED : num 0 0 0 0 0 0 0 1 1 0 ...
## $ SOMECOLL : num 0 0 0 0 0 1 0 0 0 1 ...
## $ COLLEGE : num 1 0 1 0 0 0 0 0 0 0 ...
## $ INCOME3 : num 3 1 4 4 1 3 1 0 0 4 ...
## $ INC1 : num 0 0 0 0 0 0 0 1 1 0 ...
## $ INC2 : num 0 1 0 0 1 0 1 0 0 0 ...
## $ INC3 : num 0 0 0 0 0 0 0 0 0 0 ...
## $ INC4 : num 1 0 0 0 0 1 0 0 0 0 ...
## $ DENVST4 : num 0 1 0 0 0 0 1 0 0 1 ...
## $ NODENVST : num 0 1 0 0 0 0 1 0 0 1 ...
## $ DIABETE4 : num 0 0 0 1 0 0 1 0 1 0 ...
## $ NODIABETE: num 1 1 1 0 1 1 0 1 0 1 ...
## $ SMOKER4 : num 0 0 0 1 1 0 0 0 0 0 ...
## $ FRMRSMK : num 0 0 0 1 1 0 0 0 0 0 ...
## $ SMOKE : num 0 0 0 0 0 0 0 0 0 0 ...
## $ EXERANY3 : num 0 1 1 0 0 1 0 0 1 1 ...
## $ NOEXER : num 1 0 0 1 1 0 1 1 0 0 ...
## $ BMICAT : num 1 2 1 2 2 3 3 3 3 3 ...
## $ UNDWT : num 0 0 0 0 0 0 0 0 0 0 ...
## $ OVWT : num 0 1 0 1 1 0 0 0 0 0 ...
## $ OBESE : num 0 0 0 0 0 1 1 1 1 1 ...
##
## > nrow(BRFSS)
## [1] 266109
##
## > ncol(BRFSS)
## [1] 52
##
## > colnames(BRFSS)
## [1] "CVDINFR4" "RMVTETH4" "SEX1" "X_HISPANC" "X_MRACE1"
## [6] "X_AGEG5YR" "EDUCA" "INCOME2" "X_DENVST3" "DIABETE3"
## [11] "X_SMOKER3" "EXERANY2" "X_BMI5CAT" "CVDINFR5" "RMVTETH5"
## [16] "MOSTTEETH" "FEWTEETH" "NOTEETH" "SEX2" "MALE"
## [21] "HISPANC2" "HISPANIC" "RACEGRP" "BLACK" "ASIAN"
## [26] "OTHRACE" "AGEG5YR2" "AGE1" "AGE2" "AGE3"
## [31] "EDGRP" "LOWED" "SOMECOLL" "COLLEGE" "INCOME3"
## [36] "INC1" "INC2" "INC3" "INC4" "DENVST4"
## [41] "NODENVST" "DIABETE4" "NODIABETE" "SMOKER4" "FRMRSMK"
## [46] "SMOKE" "EXERANY3" "NOEXER" "BMICAT" "UNDWT"
## [51] "OVWT" "OBESE"
##
## > data_dictionary <- describe(BRFSS)
##
## > sink("../output/data_dictionary.txt", append = TRUE)
This concluded the data cleaning, wrangling, and preparation of the data set.
Return to the Main Overview